Course
If you have little experience in applying machine learning algorithm, you would have discovered that it does not require any knowledge of Statistics as a prerequisite.
However, knowing some statistics can be beneficial to understand machine learning technically as well intuitively. Knowing some statistics will eventually be required when you want to start validating your results and interpreting them. After all, when there is data, there are statistics. Like Mathematics is the language of Science. Statistics is one of a kind language for Data Science and Machine Learning.
Statistics is a field of mathematics with lots of theories and findings. However, there are various concepts, tools, techniques, and notations are taken from this field to make machine learning what it is today. You can use descriptive statistical methods to help transform observations into useful information that you will be able to understand and share with others. You can use inferential statistical techniques to reason from small samples of data to whole domains. Later in this post, you will study descriptive and inferential statistics. So, don't worry.
Before getting started, let's walk through ten examples where statistical methods are used in an applied machine learning project:
- Problem Framing: Requires the use of exploratory data analysis and data mining.
- Data Understanding: Requires the use of summary statistics and data visualization.
- Data Cleaning: Requires the use of outlier detection, imputation and more.
- Data Selection: Requires the use of data sampling and feature selection methods.
- Data Preparation: Requires the use of data transforms, scaling, encoding and much more.
- Model Evaluation: Requires experimental design and resampling methods.
- Model Configuration: Requires the use of statistical hypothesis tests and estimation statistics.
- Model Selection: Requires the use of statistical hypothesis tests and estimation statistics.
- Model Presentation: Requires the use of estimation statistics such as confidence intervals.
- Model Predictions: Requires the use of estimation statistics such as prediction intervals.
Source: Statistical Methods for Machine Learning
Isn't that fascinating?
This post will give you a solid background in the essential but necessary statistics required for becoming a good machine learning practitioner.
In this post, you will study:
- Introduction to Statistics and its types
- Statistics for data preparation
- Statistics for model evaluation
- Gaussian and Descriptive stats
- Variable correlation
- Non-parametric Statistics
You have a lot to cover, and all of the topics are equally important. Let's get started!
Introduction to Statistics and its Types
Let's briefly study how to define statistics in simple terms.
Statistics is considered a subfield of mathematics. It refers to a multitude of methods for working with data and using that data to answer many types of questions.
When it comes to the statistical tools that are used in practice, it can be helpful to divide the field of statistics into two broad groups of methods: descriptive statistics for summarizing data, and inferential statistics for concluding samples of data (Statistics for Machine Learning (7-Day Mini-Course).
- Descriptive Statistics: Descriptive statistics are used to describe the essential features of the data in a study. They provide simple summaries about the sample and the measures. Together with simple graphics analysis, they form the basis of virtually every quantitative analysis of data. The below infographic provides a good summary of descriptive statistics:

Source: IntellSpot
-
Inferential Statistics: Inferential statistics are methods that help in quantifying properties of the domain or population from a tinier set of obtained observations called a sample. Below is an infographic which beautifully describes inferential statistics:

Source: Analytics Vidhya
In the next section, you will study the use of statistics for data preparation.
Statistics for Data Preparation
Statistical methods are required in the development of train and test data for your machine learning model.
This includes techniques for:
- Outlier detection
- Missing value imputation
- Data sampling
- Data scaling
- Variable encoding
A basic understanding of data distributions, descriptive statistics, and data visualization is required to help you identify the methods to choose when performing these tasks.
Let's analyze each of the above points briefly.
Outlier detection:
Let's first see what an outlier is.
An outlier is considered an observation that appears to deviate from other observations in the sample. The following figure makes the definition more prominent.

Source: MathWorks
You can spot the outliers in the data as given the above figure.
Many machine learning algorithms are sensitive to the range and distribution of attribute values in the input data. Outliers in input data can skew and mislead the training process of machine learning algorithms resulting in longer training times, less accurate models and ultimately more mediocre results.
Identification of potential outliers is vital for the following reasons:
-
An outlier could indicate the data is bad. In example, the data maybe coded incorrectly, or the experiment did not run correctly. If it can be determined that an outlying point is, in fact, erroneous, then the value that is outlying should be removed from the analysis. If it is possible to correct that is another option.
-
In a few cases, it may not be possible to determine whether an outlying point is a bad data point. Outliers could be due to random variation or could possibly indicate something scientifically interesting. In any event, you typically do not want to just delete the outlying observation. However, if the data contains significant outliers, you may need to consider the use of robust statistical techniques.
So, outliers are often not good for your predictive models (Although, sometimes, these outliers can be used as an advantage. But that is out of the scope of this post). You need the statistical know-how to handle outliers efficiently.
Missing value imputation:
Well, most of the datasets now suffer from the problem of missing values. Your machine learning model may not get trained effectively if the data that you are feeding to the model contains missing values. Statistical tools and techniques come here for the rescue.
Many people tend to discard the data instances which contain a missing value. But that is not a good practice because during that course you may lose essential features/representations of the data. Although there are advanced methods for dealing with missing value problems, these are the quick techniques that one would go for: Mean Imputation and Median Imputation.
It is imperative that you understand what mean and median are.
Say, you have a feature X1 which has these values - 13, 18, 13, 14, 13, 16, 14, 21, 13
The mean is the usual average, so I'll add and then divide:
(13 + 18 + 13 + 14 + 13 + 16 + 14 + 21 + 13) / 9 = 15
Note that the mean, in this case, isn't a value from the original list. This is a common result. You should not assume that your mean will be one of your original numbers.
The median is the middle value, so first, you will have to rewrite the list in numerical order:
13, 13, 13, 13, 14, 14, 16, 18, 21
There are nine numbers in the list, so the middle one will be the (9 + 1) / 2 = 10 / 2 = 5th number:
13, 13, 13, 13, 14, 14, 16, 18, 21
So the median is 14.
Data sampling:
Data is considered the currency of applied machine learning. Therefore, its collection and usage both are equally significant.
Data sampling refers to statistical methods for selecting observations from the domain with the objective of estimating a population parameter. In other words, sampling is an active process of gathering observations with the intent of estimating a population variable.
Each row of a dataset represents an observation that is indicative of a particular population. When working with data, you often do not have access to all possible observations. This could be for many reasons, for example:
- It may be difficult or expensive to make more observations.
- It may be challenging to gather all the observations together.
- More observations are expected to be made in the future.
Many times, you will not have the right proportion of the data samples. So, you will have to under-sample or over-sample based on the type of problem.
You perform under-sampling when the data samples for a particular category are very high compared to other meaning you discard some of the data samples from the category where they are higher. You perform over-sampling when the data samples for a particular type are decidedly lower compared to the other. In this case, you generate data samples.
This applies to multi-class scenarios as well.
Statistical sampling is a large field of study, but in applied machine learning, there may be three types of sampling that you are likely to use: simple random sampling, systematic sampling, and stratified sampling.
- Simple Random Sampling: Samples are drawn with a uniform probability from the domain.
- Systematic Sampling: Samples are drawn using a pre-specified pattern, such as at intervals.
- Stratified Sampling: Samples are drawn within pre-specified categories (i.e., strata).
Although these are the more common types of sampling that you may encounter, there are other techniques (A Gentle Introduction to Statistical Sampling and Resampling).
Data Scaling:
Often, the features of your dataset may widely vary in ranges. Some features may have a scale of 0 to 100 while the other may have ranges of 0.01 - 0.001, 10000- 20000, etc.
This is very problematic for efficient modeling. Because a small change in the feature which has a lower value range than the other feature may not have a significant impact on those other features. It affects the process of good learning. Dealing with this problem is known as data scaling.
There are different data scaling techniques such as Min-Max scaling, Absolute scaling, Standard scaling, etc.
Variable encoding:
At times, your datasets contain a mixture of both numeric and non-numeric data. Many machine learning frameworks like scikit-learn expect all the data to be present in all numeric format. This is also helpful to speed up the computation process.
Again, statistics come for saving you.
Techniques like Label encoding, One-Hot encoding, etc. are used to convert non-numeric data to numeric.
It's Time to Apply the Techniques!
You have covered a lot of theory for now. You will apply some of these to get the real feel.
You will start off by applying some statistical methods to detect Outliers.
You will use the Z-Score index to detect outliers, and for this, you will investigate the Boston House Price dataset. Let's start off by importing the dataset from sklearn's utilities, and as you go along, you will start the necessary concepts.
import pandas as pd
import numpy as np
from sklearn.datasets import load_boston
# Load the Boston dataset into a variable called boston
boston = load_boston()
# Separate the features from the target
x = boston.data
y = boston.target
To view the dataset in a standard tabular format with the all the feature names, you will convert this into a pandas dataframe.
# Take the columns separately in a variable
columns = boston.feature_names
# Create the dataframe
boston_df = pd.DataFrame(boston.data)
boston_df.columns = columns
boston_df.head()
It is a common practice to start with univariate outlier analysis where you consider just one feature at a time. Often, a simple box-plot of a particular feature can give you good starting point. You will make a box-plot using seaborn and you will use the DIS feature.
import seaborn as sns
sns.boxplot(x=boston_df['DIS'])
import matplotlib.pyplot as plt
plt.show()
<matplotlib.axes._subplots.AxesSubplot at 0x8abded0>
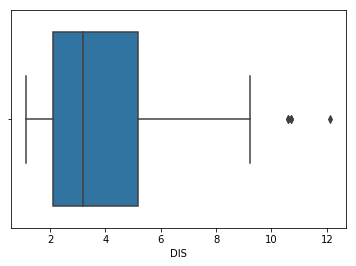
To view the box-plot, you did the second import of matplotlib since seaborn plots are displayed like ordinary matplotlib plots.
The above plot shows three points between 10 to 12, these are outliers as they're are not included in the box of other observations. Here you analyzed univariate outlier, i.e., you used DIS feature only to check for the outliers.
Let's proceed with Z-Score now.
"The Z-score is the signed number of standard deviations by which the value of an observation or data point is above the mean value of what is being observed or measured." - Wikipedia
The idea behind Z-score is to describe any data point regarding their relationship with the Standard Deviation and Mean for the group of data points. Z-score is about finding the distribution of data where the mean is 0, and the standard deviation is 1, i.e., normal distribution.
Wait! How on earth does this help in identifying the outliers?
Well, while calculating the Z-score you re-scale and center the data (mean of 0 and standard deviation of 1) and look for the instances which are too far from zero. These data points that are way too far from zero are treated as the outliers. In most common cases the threshold of 3 or -3 is used. In example, say the Z-score value is greater than or less than 3 or -3 respectively. This data point will then be identified as an outlier.
You will use the Z-score function defined in scipy library to detect the outliers.
from scipy import stats
z = np.abs(stats.zscore(boston_df))
print(z)
[[0.41771335 0.28482986 1.2879095 ... 1.45900038 0.44105193 1.0755623 ]
[0.41526932 0.48772236 0.59338101 ... 0.30309415 0.44105193 0.49243937]
[0.41527165 0.48772236 0.59338101 ... 0.30309415 0.39642699 1.2087274 ]
...
[0.41137448 0.48772236 0.11573841 ... 1.17646583 0.44105193 0.98304761]
[0.40568883 0.48772236 0.11573841 ... 1.17646583 0.4032249 0.86530163]
[0.41292893 0.48772236 0.11573841 ... 1.17646583 0.44105193 0.66905833]]
It is not possible to detect the outliers by just looking at the above output. You are more intelligent! You will define the threshold for yourself, and you will use a simple condition for detecting the outliers that cross your threshold.
threshold = 3
print(np.where(z > 3))
(array([ 55, 56, 57, 102, 141, 142, 152, 154, 155, 160, 162, 163, 199,
200, 201, 202, 203, 204, 208, 209, 210, 211, 212, 216, 218, 219,
220, 221, 222, 225, 234, 236, 256, 257, 262, 269, 273, 274, 276,
277, 282, 283, 283, 284, 347, 351, 352, 353, 353, 354, 355, 356,
357, 358, 363, 364, 364, 365, 367, 369, 370, 372, 373, 374, 374,
380, 398, 404, 405, 406, 410, 410, 411, 412, 412, 414, 414, 415,
416, 418, 418, 419, 423, 424, 425, 426, 427, 427, 429, 431, 436,
437, 438, 445, 450, 454, 455, 456, 457, 466], dtype=int32), array([ 1, 1, 1, 11, 12, 3, 3, 3, 3, 3, 3, 3, 1, 1, 1, 1, 1,
1, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 5, 3, 3, 1, 5,
5, 3, 3, 3, 3, 3, 3, 1, 3, 1, 1, 7, 7, 1, 7, 7, 7,
3, 3, 3, 3, 3, 5, 5, 5, 3, 3, 3, 12, 5, 12, 0, 0, 0,
0, 5, 0, 11, 11, 11, 12, 0, 12, 11, 11, 0, 11, 11, 11, 11, 11,
11, 0, 11, 11, 11, 11, 11, 11, 11, 11, 11, 11, 11, 11, 11],
dtype=int32))
Again, a confusing output! The first array contains the list of row numbers and the second array contains their respective column numbers. For example, z[55][1] have a Z-score higher than 3.
print(z[55][1])
3.375038763517309
So, the 55th record on column ZN is an outlier. You can extend things from here.
You saw how you could use Z-Score and set its threshold to detect potential outliers in the data. Next, you will see how to do some missing value imputation.
You will use the famous Pima Indian Diabetes dataset which is known to have missing values. But before proceeding any further, you will have to load the dataset into your workspace.
You will load the dataset into a DataFrame object data.
data = pd.read_csv("https://raw.githubusercontent.com/jbrownlee/Datasets/master/pima-indians-diabetes.data.csv",header=None)
print(data.describe())
0 1 2 3 4 5 \
count 768.000000 768.000000 768.000000 768.000000 768.000000 768.000000
mean 3.845052 120.894531 69.105469 20.536458 79.799479 31.992578
std 3.369578 31.972618 19.355807 15.952218 115.244002 7.884160
min 0.000000 0.000000 0.000000 0.000000 0.000000 0.000000
25% 1.000000 99.000000 62.000000 0.000000 0.000000 27.300000
50% 3.000000 117.000000 72.000000 23.000000 30.500000 32.000000
75% 6.000000 140.250000 80.000000 32.000000 127.250000 36.600000
max 17.000000 199.000000 122.000000 99.000000 846.000000 67.100000
6 7 8
count 768.000000 768.000000 768.000000
mean 0.471876 33.240885 0.348958
std 0.331329 11.760232 0.476951
min 0.078000 21.000000 0.000000
25% 0.243750 24.000000 0.000000
50% 0.372500 29.000000 0.000000
75% 0.626250 41.000000 1.000000
max 2.420000 81.000000 1.000000
You might have already noticed that the column names are numeric here. This is because you are using an already preprocessed dataset. But don't worry, you will discover the names soon.
Now, this dataset is known to have missing values, but for your first glance at the above statistics, it might appear that the dataset does not contain missing values at all. But if you take a closer look, you will find that there are some columns where a zero value is entirely invalid. These are the values that are missing.
Specifically, the below columns have an invalid zero value as the minimum:
- Plasma glucose concentration
- Diastolic blood pressure
- Triceps skinfold thickness
- 2-Hour serum insulin
- Body mass index
Let's confirm this by looking at the raw data, the example prints the first 20 rows of data.
data.head(20)
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 6 | 148 | 72 | 35 | 0 | 33.6 | 0.627 | 50 | 1 |
| 1 | 1 | 85 | 66 | 29 | 0 | 26.6 | 0.351 | 31 | 0 |
| 2 | 8 | 183 | 64 | 0 | 0 | 23.3 | 0.672 | 32 | 1 |
| 3 | 1 | 89 | 66 | 23 | 94 | 28.1 | 0.167 | 21 | 0 |
| 4 | 0 | 137 | 40 | 35 | 168 | 43.1 | 2.288 | 33 | 1 |
| 5 | 5 | 116 | 74 | 0 | 0 | 25.6 | 0.201 | 30 | 0 |
| 6 | 3 | 78 | 50 | 32 | 88 | 31.0 | 0.248 | 26 | 1 |
| 7 | 10 | 115 | 0 | 0 | 0 | 35.3 | 0.134 | 29 | 0 |
| 8 | 2 | 197 | 70 | 45 | 543 | 30.5 | 0.158 | 53 | 1 |
| 9 | 8 | 125 | 96 | 0 | 0 | 0.0 | 0.232 | 54 | 1 |
| 10 | 4 | 110 | 92 | 0 | 0 | 37.6 | 0.191 | 30 | 0 |
| 11 | 10 | 168 | 74 | 0 | 0 | 38.0 | 0.537 | 34 | 1 |
| 12 | 10 | 139 | 80 | 0 | 0 | 27.1 | 1.441 | 57 | 0 |
| 13 | 1 | 189 | 60 | 23 | 846 | 30.1 | 0.398 | 59 | 1 |
| 14 | 5 | 166 | 72 | 19 | 175 | 25.8 | 0.587 | 51 | 1 |
| 15 | 7 | 100 | 0 | 0 | 0 | 30.0 | 0.484 | 32 | 1 |
| 16 | 0 | 118 | 84 | 47 | 230 | 45.8 | 0.551 | 31 | 1 |
| 17 | 7 | 107 | 74 | 0 | 0 | 29.6 | 0.254 | 31 | 1 |
| 18 | 1 | 103 | 30 | 38 | 83 | 43.3 | 0.183 | 33 | 0 |
| 19 | 1 | 115 | 70 | 30 | 96 | 34.6 | 0.529 | 32 | 1 |
Clearly there are 0 values in the columns 2, 3, 4, and 5.
As this dataset has missing values denoted as 0, so it might be tricky to handle it by just using the conventional means. Let's summarize the approach you will follow to combat this:
- Get the count of zeros in each of the columns you saw earlier.
- Determine which columns have the most zero values from the previous step.
- Replace the zero values in those columns with
NaN. - Check if the NaNs are getting appropriately reflected.
- Call the fillna() function with the imputation strategy.
# Step 1: Get the count of zeros in each of the columns
print((data[[1,2,3,4,5]] == 0).sum())
1 5
2 35
3 227
4 374
5 11
dtype: int64
You can see that columns 1,2 and 5 have just a few zero values, whereas columns 3 and 4 show a lot more, nearly half of the rows.
# Step -2: Mark zero values as missing or NaN
data[[1,2,3,4,5]] = data[[1,2,3,4,5]].replace(0, np.NaN)
# Count the number of NaN values in each column
print(data.isnull().sum())
0 0
1 5
2 35
3 227
4 374
5 11
6 0
7 0
8 0
dtype: int64
Let's get sure at this point of time that your NaN replacement was a hit by taking a look at the dataset as a whole:
# Step 4
data.head(20)
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 6 | 148.0 | 72.0 | 35.0 | NaN | 33.6 | 0.627 | 50 | 1 |
| 1 | 1 | 85.0 | 66.0 | 29.0 | NaN | 26.6 | 0.351 | 31 | 0 |
| 2 | 8 | 183.0 | 64.0 | NaN | NaN | 23.3 | 0.672 | 32 | 1 |
| 3 | 1 | 89.0 | 66.0 | 23.0 | 94.0 | 28.1 | 0.167 | 21 | 0 |
| 4 | 0 | 137.0 | 40.0 | 35.0 | 168.0 | 43.1 | 2.288 | 33 | 1 |
| 5 | 5 | 116.0 | 74.0 | NaN | NaN | 25.6 | 0.201 | 30 | 0 |
| 6 | 3 | 78.0 | 50.0 | 32.0 | 88.0 | 31.0 | 0.248 | 26 | 1 |
| 7 | 10 | 115.0 | NaN | NaN | NaN | 35.3 | 0.134 | 29 | 0 |
| 8 | 2 | 197.0 | 70.0 | 45.0 | 543.0 | 30.5 | 0.158 | 53 | 1 |
| 9 | 8 | 125.0 | 96.0 | NaN | NaN | NaN | 0.232 | 54 | 1 |
| 10 | 4 | 110.0 | 92.0 | NaN | NaN | 37.6 | 0.191 | 30 | 0 |
| 11 | 10 | 168.0 | 74.0 | NaN | NaN | 38.0 | 0.537 | 34 | 1 |
| 12 | 10 | 139.0 | 80.0 | NaN | NaN | 27.1 | 1.441 | 57 | 0 |
| 13 | 1 | 189.0 | 60.0 | 23.0 | 846.0 | 30.1 | 0.398 | 59 | 1 |
| 14 | 5 | 166.0 | 72.0 | 19.0 | 175.0 | 25.8 | 0.587 | 51 | 1 |
| 15 | 7 | 100.0 | NaN | NaN | NaN | 30.0 | 0.484 | 32 | 1 |
| 16 | 0 | 118.0 | 84.0 | 47.0 | 230.0 | 45.8 | 0.551 | 31 | 1 |
| 17 | 7 | 107.0 | 74.0 | NaN | NaN | 29.6 | 0.254 | 31 | 1 |
| 18 | 1 | 103.0 | 30.0 | 38.0 | 83.0 | 43.3 | 0.183 | 33 | 0 |
| 19 | 1 | 115.0 | 70.0 | 30.0 | 96.0 | 34.6 | 0.529 | 32 | 1 |
You can see that marking the missing values had the intended effect.
Up till now, you analyzed essential trends when data is missing and how you can make use of simple statistical measures to get a hold of it. Now, you will impute the missing values using Mean Imputation which is essentially imputing the mean of the respective column in place of missing values.
# Step 5: Call the fillna() function with the imputation strategy
data.fillna(data.mean(), inplace=True)
# Count the number of NaN values in each column to verify
print(data.isnull().sum())
0 0
1 0
2 0
3 0
4 0
5 0
6 0
7 0
8 0
dtype: int64
Excellent!
This DataCamp article effectively guides you in implementing data scaling as a data preprocessing step. Be sure to check it out.
Next, you will do variable encoding.
Before that, you need a dataset which actually contains non-numeric data. You will use the famous Iris dataset for this.
# Load the dataset to a DataFrame object iris
iris = pd.read_csv("http://archive.ics.uci.edu/ml/machine-learning-databases/iris/iris.data",header=None)
# See first 20 rows of the dataset
iris.head(20)
| 0 | 1 | 2 | 3 | 4 | |
|---|---|---|---|---|---|
| 0 | 5.1 | 3.5 | 1.4 | 0.2 | Iris-setosa |
| 1 | 4.9 | 3.0 | 1.4 | 0.2 | Iris-setosa |
| 2 | 4.7 | 3.2 | 1.3 | 0.2 | Iris-setosa |
| 3 | 4.6 | 3.1 | 1.5 | 0.2 | Iris-setosa |
| 4 | 5.0 | 3.6 | 1.4 | 0.2 | Iris-setosa |
| 5 | 5.4 | 3.9 | 1.7 | 0.4 | Iris-setosa |
| 6 | 4.6 | 3.4 | 1.4 | 0.3 | Iris-setosa |
| 7 | 5.0 | 3.4 | 1.5 | 0.2 | Iris-setosa |
| 8 | 4.4 | 2.9 | 1.4 | 0.2 | Iris-setosa |
| 9 | 4.9 | 3.1 | 1.5 | 0.1 | Iris-setosa |
| 10 | 5.4 | 3.7 | 1.5 | 0.2 | Iris-setosa |
| 11 | 4.8 | 3.4 | 1.6 | 0.2 | Iris-setosa |
| 12 | 4.8 | 3.0 | 1.4 | 0.1 | Iris-setosa |
| 13 | 4.3 | 3.0 | 1.1 | 0.1 | Iris-setosa |
| 14 | 5.8 | 4.0 | 1.2 | 0.2 | Iris-setosa |
| 15 | 5.7 | 4.4 | 1.5 | 0.4 | Iris-setosa |
| 16 | 5.4 | 3.9 | 1.3 | 0.4 | Iris-setosa |
| 17 | 5.1 | 3.5 | 1.4 | 0.3 | Iris-setosa |
| 18 | 5.7 | 3.8 | 1.7 | 0.3 | Iris-setosa |
| 19 | 5.1 | 3.8 | 1.5 | 0.3 | Iris-setosa |
You can easily convert the string values to integer values using the LabelEncoder. The three class values (Iris-setosa, Iris-versicolor, Iris-virginica) are mapped to the integer values (0, 1, 2).
In this case, the fourth column/feature of the dataset contains non-numeric values. So you need to separate it out.
# Convert the DataFrame to a NumPy array
iris = iris.values
# Separate
Y = iris[:,4]
# Label Encode string class values as integers
from sklearn.preprocessing import LabelEncoder
label_encoder = LabelEncoder()
label_encoder = label_encoder.fit(Y)
label_encoded_y = label_encoder.transform(Y)
Now, let's study another area where the need for elementary knowledge of statistics is very crucial.
Statistics for Model Evaluation
You have designed and developed your machine learning model. Now, you want to evaluate the performance of your model on the test data. In this regards, you seek help of various statistical metrics like Precision, Recall, ROC, AUC, RMSE, etc. You also seek help from multiple data resampling techniques such as k-fold Cross-Validation.
Statistics can effectively be used to:
- Estimate a hypothesis accuracy
- Determine the error of two hypotheses
- Compare learning algorithms using McNemar's test
It is important to note that the hypothesis refers to learned models; the results of running a learning algorithm on a dataset. Evaluating and comparing the hypothesis means comparing learned models, which is different from evaluating and comparing machine learning algorithms, which could be trained on different samples from the same problem or various problems.
Let's study Gaussian and Descriptive statistics now.
Introduction to Gaussian and Descriptive Stats
A sample of data is nothing but a snapshot from a broader population of all the potential observations that could be taken from a domain or generated by a process.
Interestingly, many observations fit a typical pattern or distribution called the normal distribution, or more formally, the Gaussian distribution. This is the bell-shaped distribution that you may be aware of. The following figure denotes a Gaussian distribution:
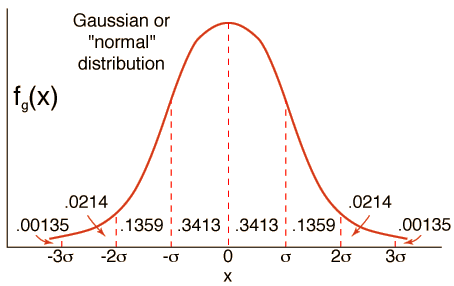
Source: HyperPhysics
Gaussian processes and Gaussian distributions are whole another sub-fields unto themselves. But, you will now study two of the most essential ingredients that build the entire world of Gaussian distributions in general.
Any sample data taken from a Gaussian distribution can be summarized with two parameters:
- Mean: The central tendency or most likely value in the distribution (the top of the bell).
- Variance: The average difference that observations have from the mean value in the distribution (the spread).
The term variance also gives rise to another critical term, i.e., standard deviation, which is merely the square root of the variance.
The mean, variance, and standard deviation can be directly calculated from data samples using numpy.
You will first generate a sample of 100 random numbers pulled from a Gaussian distribution with a mean of 50 and a standard deviation of 5. You will then calculate the summary statistics.
First, you will import all the dependencies.
# Dependencies
from numpy.random import seed
from numpy.random import randn
from numpy import mean
from numpy import var
from numpy import std
Next, you set the random number generator seed so that your results are reproducible.
seed(1)
# Generate univariate observations
data = 5 * randn(10000) + 50
# Calculate statistics
print('Mean: %.3f' % mean(data))
print('Variance: %.3f' % var(data))
print('Standard Deviation: %.3f' % std(data))
Mean: 50.049
Variance: 24.939
Standard Deviation: 4.994
Close enough, eh?
Let's study the next topic now.
Variable Correlation
Generally, the features that are contained in a dataset can often be related to each other which is very obvious to happen in practice. In statistical terms, this relationship between the features of your dataset (be it simple or complex) is often termed as correlation.
It is crucial to find out the degree of the correlation of the features in a dataset. This step essentially serves you as feature selection which concerns selecting the most important features from a dataset. This step is one of the most vital steps in a standard machine learning pipeline as it can give you a tremendous accuracy boost that too within a lesser amount of time.
For better understanding and to keep it more practical let's understand why features can be related to each other:
- One feature can be a determinant of another feature
- One feature could be associated with another feature in some degree of composition
- Multiple features can combine and give birth to another feature
Correlation between the features can be of three types: - Positive correlation where both the feature change in the same direction, Neutral correlation when there is no relationship of the change in the two features, Negative correlation where both the features change in opposite directions.
Correlation measurements form the fundamental of filter-based feature selection techniques. Check this article if you want to study more about feature selection.
You can mathematically the relationship between samples of two variables using a statistical method called Pearson’s correlation coefficient, named after the developer of the method, Karl Pearson.
You can calculate the Pearson's correlation score by using the corr() function of pandas with the method parameter as pearson. Let's study the correlation between the features of the Pima Indians Diabetes dataset that you used earlier. You already have the data in good shape.
# Data
data.head()
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 6 | 148.0 | 72.0 | 35.00000 | 155.548223 | 33.6 | 0.627 | 50 | 1 |
| 1 | 1 | 85.0 | 66.0 | 29.00000 | 155.548223 | 26.6 | 0.351 | 31 | 0 |
| 2 | 8 | 183.0 | 64.0 | 29.15342 | 155.548223 | 23.3 | 0.672 | 32 | 1 |
| 3 | 1 | 89.0 | 66.0 | 23.00000 | 94.000000 | 28.1 | 0.167 | 21 | 0 |
| 4 | 0 | 137.0 | 40.0 | 35.00000 | 168.000000 | 43.1 | 2.288 | 33 | 1 |
# Create the matrix of correlation score between the features and the label
scoreTable = data.corr(method='pearson')
# Visulaize the matrix
data.corr(method='pearson').style.format("{:.2}").background_gradient(cmap=plt.get_cmap('coolwarm'), axis=1)
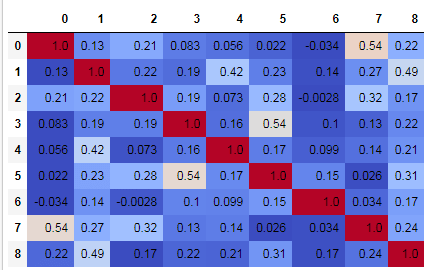
You can clearly see the Pearson's correlation between all the features and the label of the dataset.
In the next section, you will study non-parametric statistics.
Non-parametric Statistics
A large portion of the field of statistics and statistical methods is dedicated to data where the distribution is known.
Non-parametric statistics comes in handy when there is no or few information available about the population parameters. Non-parametric tests make no assumptions about the distribution of data.
In the case where you are working with nonparametric data, specialized nonparametric statistical methods can be used that discard all information about the distribution. As such, these methods are often referred to as distribution-free methods.
Bu before a nonparametric statistical method can be applied, the data must be converted into a rank format. Statistical methods that expect data in a rank format are sometimes called rank statistics. Examples of rank statistics can be rank correlation and rank statistical hypothesis tests. Ranking data is exactly as its name suggests.
A widely used nonparametric statistical hypothesis test for checking for a difference between two independent samples is the Mann-Whitney U test, named for Henry Mann and Donald Whitney.
You will implement this test in Python via the mannwhitneyu() which is provided by SciPy.
# The dependencies that you need
from scipy.stats import mannwhitneyu
from numpy.random import rand
# seed the random number generator
seed(1)
# Generate two independent samples
data1 = 50 + (rand(100) * 10)
data2 = 51 + (rand(100) * 10)
# Compare samples
stat, p = mannwhitneyu(data1, data2)
print('Statistics = %.3f, p = %.3f' % (stat, p))
# Interpret
alpha = 0.05
if p > alpha:
print('Same distribution (fail to reject H0)')
else:
print('Different distribution (reject H0)')
Statistics = 4077.000, p = 0.012
Different distribution (reject H0)
alpha is the threshold parameter which is decided by you. The mannwhitneyu() returns two things:
-
statistic: The Mann-Whitney U statistic, equal to min(U for x, U for y) if alternative is equal to None (deprecated; exists for backward compatibility), and U for y otherwise.
-
pvalue: p-value assuming an asymptotic normal distribution.
If you want to study the other methods of Non-parametric statistics, you can do it from here.
The other two popular non-parametric statistical significance tests that you can use are:
That calls for a wrap up!
You have finally made it to the end. In this article, you studied a variety of essential statistical concepts that play very crucial role in your machine learning projects. So, understanding them is just important.
From mere an introduction to statistics, you took it to statistical rankings that too with several implementations. That is definitely quite a feat. You studied three different datasets, exploited pandas and numpy functionalities to the fullest and moreover, you used SciPy as well. Next are some links for you if you want to take things further:
Following are the resources I took help from for writing this blog:
- Machine Learning Mastery mini course on Statistics
- A Gentle Introduction to Statistical Sampling and Resampling
- https://www.khanacademy.org/math/statistics-probability
- Statistical Learning course by Stanford University
Let me know your views/queries in the comments section. Also, check out DataCamp's course on "Statistical Thinking in Python" which is very practically aligned.

