Introduction
Pandas is an extraordinarily powerful tool in Python's data science ecosystem, offering several data manipulation and cleaning capabilities. However, while great for medium-sized datasets, it can face performance issues when dealing with large datasets, prompting the need for high-performance alternatives.
This comprehensive article introduces some of these alternatives and compares them through benchmarking in terms of data loading time, execution time, memory usage, scalability, and ease of use.
Note: Benchmarking results vary a lot depending on your hardware setup.
Understanding the Benchmarks
Benchmarks are a point of reference against which software or hardware may be compared for performance evaluation. This reference is relevant in software performance optimization as it allows us to measure the efficiency of different methods, algorithms, or tools. In this context, the key metrics for benchmarking data manipulation libraries include execution time, memory usage, scalability, and ease of use.
Introduction to High-Performance Alternatives
The main alternative Python tools are Polars, Vaex, and Datatable. Before diving into their comparative analysis, let’s have a quick overview of what each one of these tools is.
Polars
Polars can be described by three key features. First, it offers a comprehensive Python API filled with numerous functions to handle Dataframes. Second, it can serve dual purposes - it can be employed as a library for Dataframes or as the underlying query engine for data models. Lastly, by incorporating the secure Arrow2 implementation of the Apache Arrow specification, Polars becomes a highly efficient tool for processing large amounts of data.
You can check out our introduction to Polars tutorial, as well as a comparison between pandas 2.0 and Polars.
Vaex
Vaex is for lazy, out-of-core DataFrames (similar to Pandas) to visualize and explore big tabular datasets. It can be very efficient as it delays operations until necessary (lazy evaluation), reducing memory usage and time.
Datatable
Datatable can be used for performing large data processing (up to 100GB) on a single-node machine at the maximum speed possible. One of the important features of Datatable is its interoperability with Pandas/NumPy/pure Python, which allows users to easily convert to another data-processing framework.
To learn more about pandas, our pandas Tutorial Dataframes in Python is a great starting point.
Also, the pandas Cheat Sheet for Data Science in Python gives a quick guide to the basics of the Python data analysis library, along with code samples.
Setting up the Benchmarking Environment
This section covers the creation of the benchmarking data and the installation of all the libraries involved in the benchmarking analysis.
Benchmarking data
The benchmarking data set is 5.7 GB large, which is large enough to perform a decent comparison. Each row has been duplicated 100000 times, resulting in 76 800 000 rows. Diabetes data is the original one being used and is freely available from Kaggle.
The final benchmarking notebook is available on GitHub.
import pandas as pd
data_URL = "https://raw.githubusercontent.com/keitazoumana/Experimentation-Data/main/diabetes.csv"
original_data = pd.read_csv(data_URL)
# Duplicate each row 100000 times
benchmarking_df = original_data.loc[original_data.index.repeat(100000)]
# Save the result as a CSV file for future use.
file_name = "benchmarking_data.csv"
benchmarking_df.to_csv(file_name, index=False)Installation of the libraries
Now, we can proceed with the installation of all the libraries involved in the benchmarking analysis.
From a Jupyter notebook environment, all the installation is performed from the pip install command as follows:
%%bash
pip3 -q install -U polars
pip3 -q install vaex
pip -q install datatableAfter successfully running the above code block, we can import them with the import statement:
import pandas as pd
from time import time
import polars as pl
import vaex as vx
import datatable as dtFurthermore, we use the plotly library for visualization purpose.
import plotly.express as pxNow we are all set to proceed with the benchmarking analysis.
Benchmarking Execution Time
The execution time will be evaluated for the following operations: data loading, data offloading, data aggregation, and data filtering. A helper function is implemented for each task and will return a dictionary with two main keys: the library name and the execution time.
The plot metrics function is then used to plot the result in a graphical manner using the Plotly Express library. It has two parameters: the list of all the dictionaries from the previous helper functions and the title of the underlying graphic.
def plot_metrics(list_exec_time, graph_title):
df = pd.DataFrame(list_exec_time)
fig = px.bar(df, x='library', y= 'execution_time', title=graph_title)
fig.show()Data loading
The helper function read_csv_with_time has two main parameters: the file name to be read and the library name.
def read_csv_with_time(library_name, file_name):
final_time = 0
start_time = time.time()
if library_name.lower() == 'polars':
df = pl.read_csv(file_name)
elif library_name.lower() == 'pandas':
df = pd.read_csv(file_name)
elif library_name.lower() == 'vaex':
df = vx.read_csv(file_name)
elif library_name.lower() == 'datatable':
df = dt.fread(file_name)
else:
raise ValueError("Invalid library name. Must be 'polars', 'pandas', 'vaex', or 'datatable'")
end_time = time.time()
final_time = end_time - start_time
return {"library": library_name, "execution_time": final_time}The function can be applied using each of the libraries, starting with pandas.
pandas_time, pandas_df = read_csv_with_time('pandas', file_name)
polars_time, polars_df = read_csv_with_time('polars', file_name)
vaex_time, vaex_df = read_csv_with_time('vaex', file_name)
datatable_time, dt_df = read_csv_with_time('datatable', file_name)The resulting dictionaries can be combined as a list and plotted using the following expression.
exec_times = [pandas_time, polars_time,
vaex_time, datatable_time]
def plot_metrics(list_exec_time, graph_title):
[print(exec_time) for exec_time in list_exec_time]
df = pd.DataFrame(exec_times)
# Plot bar plot using Plotly Express
fig = px.bar(df, x='library', y='execution_time', title=graph_title)
fig.show()
plot_metrics(exec_times, "Read Execution Time Comparison"){'library': 'pandas', 'execution_time': 35.07908916473389}
{'library': 'polars', 'execution_time': 6.041005373001099}
{'library': 'vaex', 'execution_time': 36.431522607803345}
{'library': 'datatable', 'execution_time': 4.4584901332855225}Dictionary format of the execution times for data loading
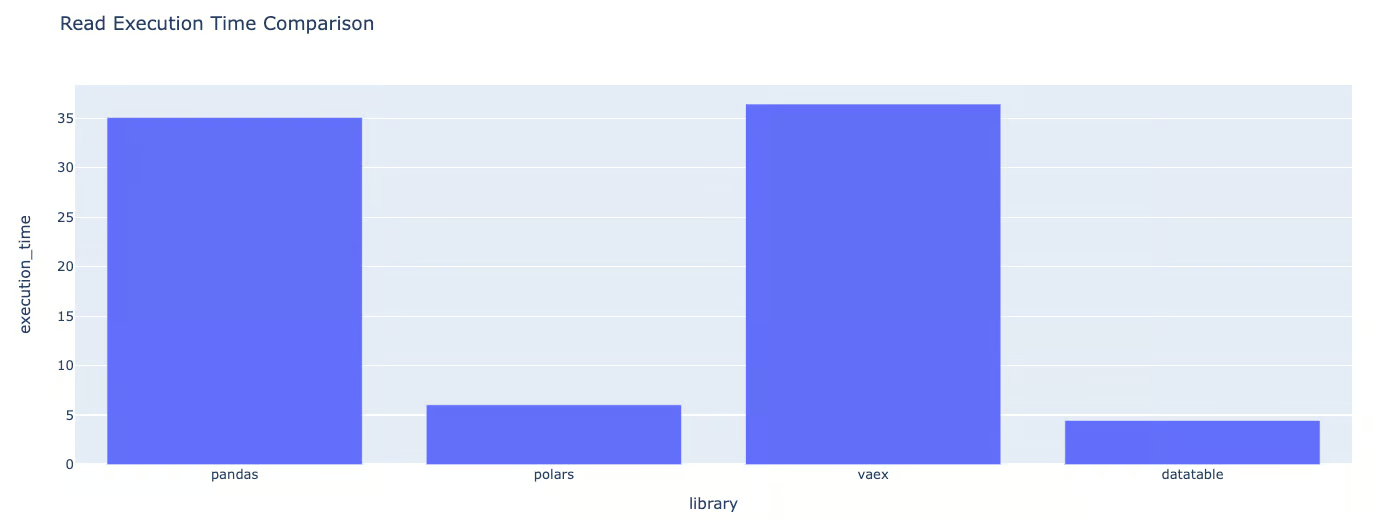
Graphic format of the execution times for data loading
Based on the graphics, we can conclude that Polars is:
- Polars is 1.35 times slower than Datatable
- Pandas is 7.87 times slower than DataTable.
- Vaex is approximately 8.17 times slower than DataTable.
Data grouping
Similarly, the group_data_with_time is responsible for tracking the execution time for grouping the column provided in parameter. In this case, we are using the Pregnancies column.
from time import time
def group_data_with_time(library_name, df, column_name='Pregnancies'):
start_time = time()
if library_name.lower() == 'polars':
df_grouped = df.group_by(column_name).first()
elif library_name.lower() == 'vaex':
df_grouped = df.groupby(column_name, agg='first')
elif library_name.lower() == 'pandas':
df_grouped = df.groupby(column_name).first()
elif library_name.lower() == 'datatable':
df_grouped = df[:, dt.first(dt.f[:]), dt.by(column_name)]
else:
raise ValueError("Invalid library name. Must be 'polars', 'vaex', or 'datatable'")
end_time = time()
final_time = end_time - start_time
return {"library": library_name, "execution_time": final_time}pandas_time = group_data_with_time('pandas', pandas_df)
polars_time = group_data_with_time('polars', polars_df)
vaex_time = group_data_with_time('vaex', vaex_df)
datatable_time = group_data_with_time('datatable', dt_df)exec_times = [pandas_time, polars_time,
vaex_time, datatable_time]
plot_metrics(exec_times, "Grouping Execution Time Comparison"){'library': 'pandas', 'execution_time': 2.382519245147705}
{'library': 'pandas', 'execution_time'': 1.0898876190185547}
{'library': 'pandas', 'execution_time': 0.8506548404693604}
{'library': 'pandas', 'execution_time': 0.34698963165283203}
Graphic format of the execution times for data grouping
For the updated data grouping task, comparing the fastest library DataTable to the others:
We can see that DataTable is the fastest among the evaluated libraries.
- Vaex is 2.45 times slower than DataTable.
- Polars is 3.14 times slower than DataTable.
- Pandas is 6.87 times slower than DataTable.
This analysis demonstrates DataTable's superior efficiency for data grouping tasks, with Vaex being relatively close in performance. Polars and Pandas exhibit similar execution times, both significantly slower than DataTable.
Column Sorting
Using the same approach, the column sorting function sorts the given column using each library.
def sort_data_with_time(library_name, df, column_name='Pregnancies'):
start_time = time()
if library_name.lower() == 'polars':
df_sorted = df.sort(column_name)
elif library_name.lower() == 'vaex':
df_sorted = df.sort(column_name)
elif library_name.lower() == 'datatable':
df_sorted = df.sort(column_name)
elif library_name.lower() == 'pandas':
df_sorted = pd.DataFrame(df).sort_values(column_name)
else:
raise ValueError("Invalid library name. Must be 'polars', 'vaex', 'datatable', or 'pandas'")
end_time = time()
final_time = end_time - start_time
return {"library": library_name, "execution_time": final_time}pandas_time = sort_data_with_time('pandas', pandas_df)
polars_time = sort_data_with_time('polars', polars_df)
vaex_time = sort_data_with_time('vaex', vaex_df)
datatable_time = sort_data_with_time('datatable', dt_df){'library': 'pandas', 'execution_time': 6.735127687454224}
{'library': 'polars', 'execution_time': 4.119458436965942}
{'library': 'vaex', 'execution_time': 1.0371737480163574}
{'library': 'datatable', 'execution_time': 0.3971593379974365}Dictionary format of the execution times for data sorting
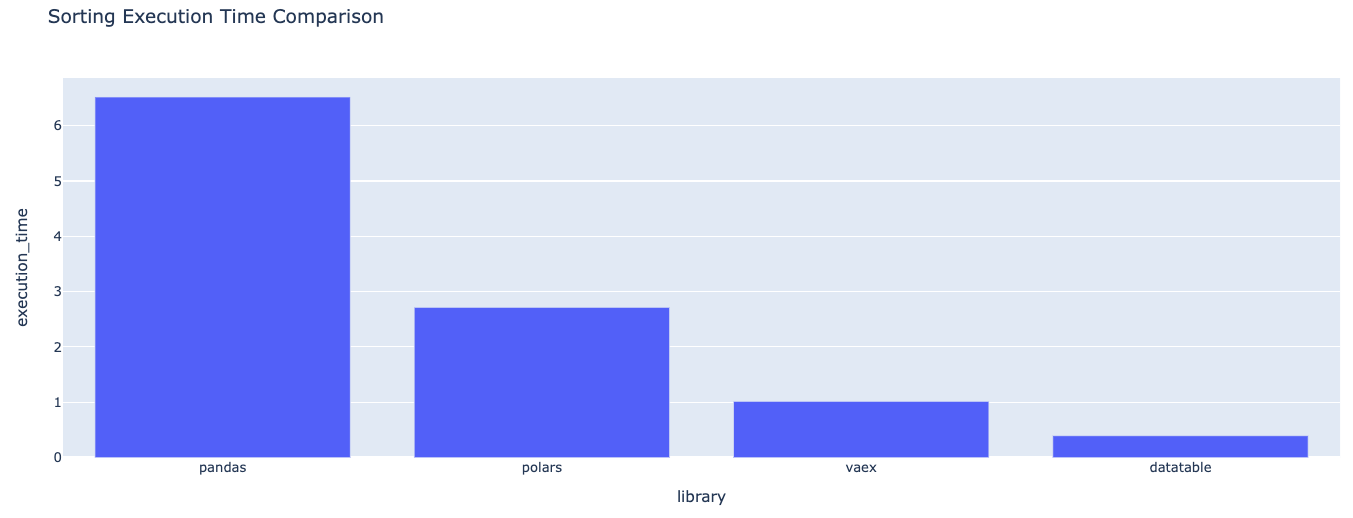
Graphic format of the execution times for data sorting
For the data sorting task, DataTable is the fastest among the evaluated libraries.
- Vaex is 2.61 times slower than DataTable.
- Polars is 10.37 times slower than DataTable.
- Pandas is 16.96 times slower than DataTable.
This analysis shows DataTable's superior performance in handling data sorting tasks, significantly outpacing Vaex, Polars, and Pandas, with Vaex being the closest in performance but still more than twice as slow.
Data Offloading
With data offloading, the idea is to convert the original data into a different format, which numpy array in this specific scenario.
def offload_data_with_time(library_name, df):
start_time = time()
if library_name.lower() == 'polars':
array = df.to_numpy()
elif library_name.lower() == 'vaex':
array = df.to_pandas_df().values
elif library_name.lower() == 'datatable':
array = df.to_numpy()
elif library_name.lower() == 'pandas':
array = pd.DataFrame(df).values
else:
raise ValueError("Invalid library name. Must be 'polars', 'vaex', 'datatable', or 'pandas'")
end_time = time()
final_time = end_time - start_time
return {"library": library_name, "execution_time": final_time}exec_times = [pandas_time, polars_time,
vaex_time, datatable_time]
plot_metrics(exec_times, "Data offloading Execution Time Comparison"){'library': 'pandas', 'execution_time': 1.8406445980072021}
{'library': 'polars', 'execution_time': 3.238591432571411}
{'library': 'vaex', 'execution_time': 6.945307970046997}
{'library': 'datatable', 'execution_time': 2.634136438369751}Dictionary format of the execution times for data offloading
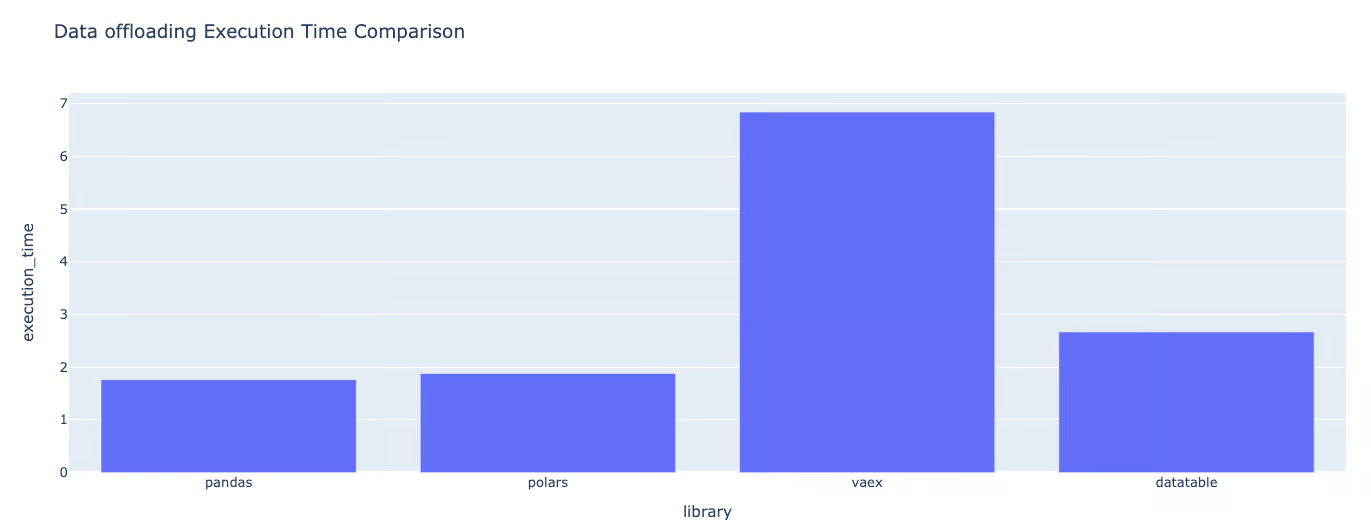
Graphic format of the execution times for data offloading
Data offloading is the last round of the benchmarking for the execution time.
This time, Vaex is the one with the highest execution time. Comparing pandas to these libraries, we can see that its execution time is approximately:
- DataTable is 1.43 times slower than Pandas.
- Polars is 1.76 times slower than Pandas.
- Vaex is 3.77 times slower than Pandas.
Benchmarking Memory Usage
The execution time is not the only criterion when comparing different libraries. Knowing how efficiently they deal with memories is also important to consider. This section shows two different memory usage using the tracemalloc library:
- Current memory usage. The total amount of memory used during the execution using a specific library. It corresponds to the RAM space being occupied at that time.
- Peak memory usage. Corresponds to the maximum amount of memory used by the program. It is important to know that the peak memory is always higher than the current memory.
Having these visualizations helps understand which program uses as much memory, leading to out-of-memory errors, especially when working with large datasets.
In addition to tracemalloc, the os library is also required.
import tracemalloc as tm
import osBefore starting, we create an empty list that will hold the result of each memory usage value.
list_memory_usage = []For each library, the estimation of the memory usage is done with the general syntax below corresponding to the execution with the Vaex library. Also, this focuses only on the memory usage for the data offloading tasks.
tm.start()
vaex_time = offload_data_with_time('vaex', vaex_df)
memory_usage = tm.get_traced_memory()
tm.stop()
list_memory_usage.append({
'library': 'vaex',
'memory_usage': memory_usage
})The syntax is the same for the remaining libraries, and we get the following instructions, respectively for Polars, pandas, and Datatable:
tm.start()
polars_time = offload_data_with_time('polars', polars_df)
memory_usage = tm.get_traced_memory()
tm.stop()
list_memory_usage.append({
'library': 'polars',
'memory_usage': memory_usage
})tm.start()
offload_data_with_time('pandas', pandas_df)
# Get the memory usage
memory_usage = tm.get_traced_memory()
tm.stop()
list_memory_usage.append({
'library': 'pandas',
'memory_usage': memory_usage
})tm.start()
datatable_time = offload_data_with_time('datatable', datatable_df)
memory_usage = tm.get_traced_memory()
tm.stop()
list_memory_usage.append({
'library': 'datatable',
'memory_usage': memory_usage
})After the execution of all the previous code, we can plot the graphic using the helper function below:
def plot_memory_usage(list_memory_usage, graph_title='Memory Usage by Library'):
df = pd.DataFrame(list_memory_usage)
# separate the memory usage tuple into two columns: current_memory and peak_memory
df[['current_memory', 'peak_memory']] = pd.DataFrame(df['memory_usage'].tolist(), index=df.index)
# now we no longer need the memory_usage column
df = df.drop(columns='memory_usage')
# melt the DataFrame to make it suitable for grouped bar chart
df_melted = df.melt(id_vars='library', var_name='memory_type', value_name='memory')
# create the grouped bar chart
fig = px.bar(df_melted, x='library', y='memory', color='memory_type', barmode='group',
labels={'memory':'Memory Usage (bytes)', 'library':'Library', 'memory_type':'Memory Type'},
title=graph_title)
fig.update_layout(yaxis_type="log")
fig.show()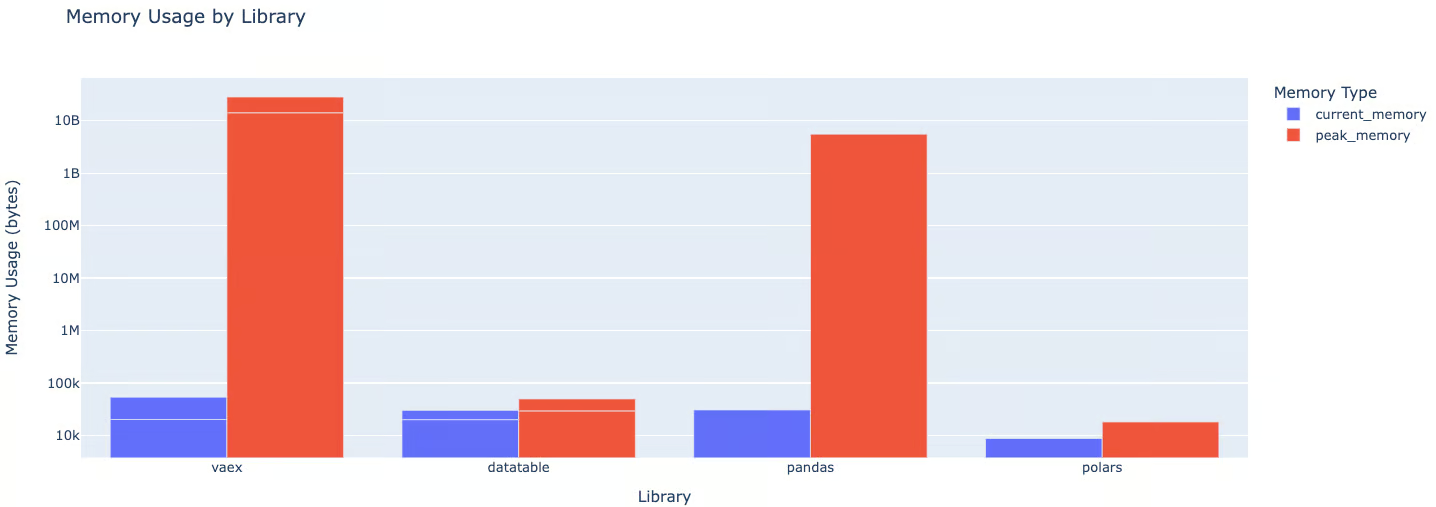
We can notice that:
- Pandas uses approximately 1100 times more current memory than Polars, 7.4 times more than Vaex, and 29.4 times more than Datatable.
- When it comes to peak memory usage, both pandas and Vaex use around 963K times more memory than Polars and 131K times more than Datatable.
Further analysis, such as Dask vs pandas speed, could provide a broader overview.
Pandas Alternatives Comparison Table
Below, we’ve compiled our findings into a comparison table, showing the differences between Polars, Vaex, and Datatables:
|
Benchmark Criteria |
Polars |
Vaex |
Datatable |
|
Data Loading Time |
Faster than Vaex, slower than Datatable |
Slower than both Polars and Datatable |
Fastest among the three |
|
Data Grouping Time |
Slowest among the three |
Faster than Polars, slower than Datatable |
Fastest among the three |
|
Data Sorting Time |
Faster than Vaex, slower than Datatable |
Slower than both Polars and Datatable |
Fastest among the three |
|
Data Offloading Time |
Faster than Vaex, slower than Datatable |
Slowest among the three |
Faster than Polars, slower than Vaex |
|
Current Memory Usage |
Uses least memory |
Uses more memory than Polars but less than Datatable |
Uses most memory among the three |
|
Peak Memory Usage |
Uses least memory |
Uses more memory than Polars but less than Datatable |
Uses most memory among the three |
|
Scalability |
Scalable with data size |
Scalable with data size, but may use more memory as data size increases |
Highly scalable with data size |
|
Ease of use |
Has a complete Python API and is compatible with Apache Arrow |
Utilizes lazy evaluation and is compatible with Pandas |
Offers interoperability with Pandas/NumPy/pure Python |
Conclusion
Different factors must be considered when performing the scalability and ease of integration analysis. Our benchmarking highlighted that even though pandas is well-established and user-friendly and has a steeper learning curve, it does not efficiently handle large datasets.
However, there are different ways that be adapted on how to speed up pandas, and our High-Performance Data Manipulation in Python: Pandas 2.0 vs Polars can help with that.
Vaex performance varies depending on the task. So, choosing an alternative to Pandas depends on the users’ specific demands, dataset sizes, and their ability to handle the learning curve and integration.

A multi-talented data scientist who enjoys sharing his knowledge and giving back to others, Zoumana is a YouTube content creator and a top tech writer on Medium. He finds joy in speaking, coding, and teaching . Zoumana holds two master’s degrees. The first one in computer science with a focus in Machine Learning from Paris, France, and the second one in Data Science from Texas Tech University in the US. His career path started as a Software Developer at Groupe OPEN in France, before moving on to IBM as a Machine Learning Consultant, where he developed end-to-end AI solutions for insurance companies. Zoumana joined Axionable, the first Sustainable AI startup based in Paris and Montreal. There, he served as a Data Scientist and implemented AI products, mostly NLP use cases, for clients from France, Montreal, Singapore, and Switzerland. Additionally, 5% of his time was dedicated to Research and Development. As of now, he is working as a Senior Data Scientist at IFC-the world Bank Group.



