Course
The DeepSeek-R1-0528 is the latest iteration of the DeepSeek R1 model, featuring enhanced reasoning and inference capabilities. It is on par with leading models like o3 and Gemini 2.5 Pro, making it the best open-source reasoning model in the world.
In this tutorial, we will fine-tune the DeepSeek-R1-0528-Qwen3-8B model on a Medical Reasoning Multiple Choice Question (MCQ) dataset. The fine-tuning process introduces a new reasoning style and answer generation in the form of A, B, C, D options.
The best part? This can be done on a consumer GPU like the RTX 4090, allowing you to fine-tune the model locally without any issues. You can check out our full guide on DeepSeek-R1 and our fine-tuning guide to learn more.
 Image by Author
Image by Author
1. Setting Up
We will set up a RunPod instance with an RTX 4090 GPU and the PyTorch 2.4.0 image.
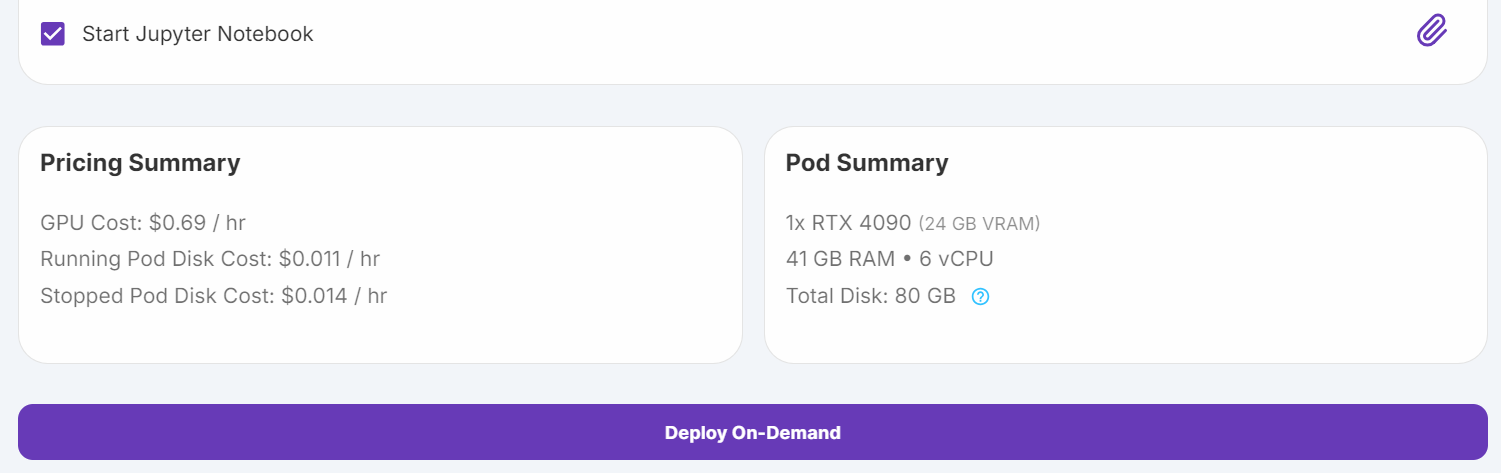
Source: My Pods
Navigate to the "My Pods" section in the RunPod console. Edit the pod configuration:
- Increase storage to 100 GB to accommodate larger datasets and models.
- Add the Hugging Face Token as an environment variable. This token will be used for authentication when interacting with Hugging Face's API.
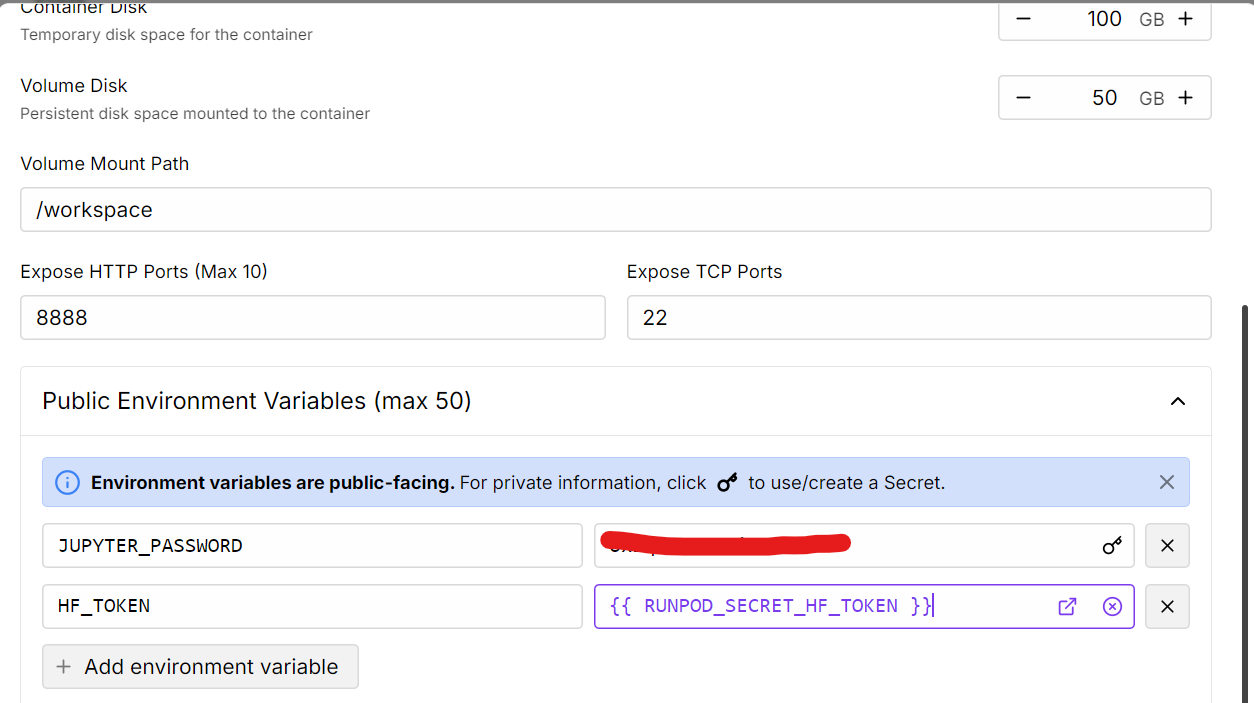
Source: My Pods
Once the pod is running, install the necessary Python packages.
Note that the latest version of the Transformers library has some known issues, so we will install an older, stable version instead.
%%capture
%pip install -U transformers==4.52.1
%pip install -U datasets
%pip install -U accelerate
%pip install -U peft
%pip install -U trl
%pip install -U bitsandbytesTo load models or push updates to the Hugging Face Hub, log in to the Hugging Face CLI using your API key
from huggingface_hub import login
import os
hf_token = os.environ.get("HF_TOKEN")
login(hf_token)2. Loading the Model and Tokenizer
Even though we have 24GB of VRAM, which is sufficient to load the full model, we will use 4-bit quantization to leave more memory available for fine-tuning and to ensure stability. This approach allows us to optimize VRAM usage while maintaining performance.
We will download and load the DeepSeek-R1-0528-Qwen3-8B model and tokenizer from Hugging Face Hub. You can learn more about working with Hugging Face and a guide to the Hub with our course.
from transformers import AutoModelForCausalLM, AutoTokenizer, BitsAndBytesConfig
import torch
bnb_config = BitsAndBytesConfig(
load_in_4bit=True,
bnb_4bit_use_double_quant=False,
bnb_4bit_quant_type="nf4",
bnb_4bit_compute_dtype=torch.bfloat16,
)
# Load tokenizer & model
model_dir = "deepseek-ai/DeepSeek-R1-0528-Qwen3-8B"
tokenizer = AutoTokenizer.from_pretrained(model_dir, use_fast=True)
model = AutoModelForCausalLM.from_pretrained(
model_dir,
quantization_config=bnb_config,
device_map="auto",
torch_dtype=torch.bfloat16,
trust_remote_code=True
)
model.config.use_cache = False
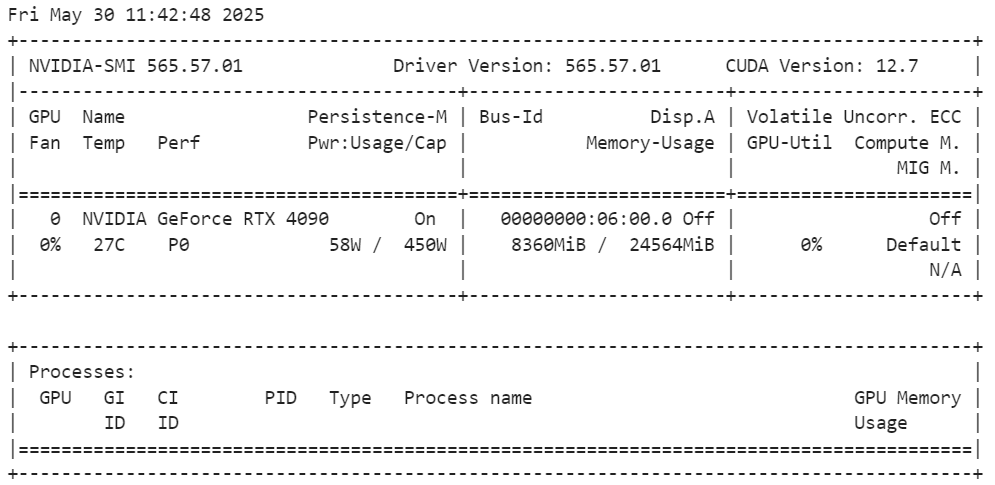
model.config.pretraining_tp = 1To verify the VRAM usage after loading the model, run the following command:
!nvidia-smiThe VRAM usage is approximately 8.3GB, leaving two-thirds of the memory available for fine-tuning.
3. Loading and Processing the Dataset
In this step, we will prepare the training prompts by creating a template with placeholders for the question and response. We will also define a Python function to format the dataset according to the prompt style and create a new “text” column.
train_prompt_style = """
Please answer with one of the options in the bracket. Write reasoning in between <analysis></analysis>. Write the answer in between <answer></answer>.
### Question:
{}
### Response:
{}"""
EOS_TOKEN = tokenizer.eos_token # Must add EOS_TOKEN
def formatting_prompts_func(examples):
inputs = examples["input"]
outputs = examples["output"]
texts = []
for question, response in zip(inputs, outputs):
# Remove the "Q:" prefix from the question
question = question.replace("Q:", "")
# Append the EOS token to the response if it's not already there
if not response.endswith(tokenizer.eos_token):
response += tokenizer.eos_token
text = train_prompt_style.format(question, response)
texts.append(text)
return {"text": texts}Then, we will load the mamachang/medical-reasoning, apply the formatting_prompts_func function, and display the “text” column from the 11th sample.
from datasets import load_dataset
dataset = load_dataset(
"mamachang/medical-reasoning",
split="train",
trust_remote_code=True,
)
dataset = dataset.map(
formatting_prompts_func,
batched=True,
)
print(dataset["text"][10])The text column contains the system prompt, question, reasoning, and answer in the following format:
Please answer with one of the options in the bracket. Write reasoning in between <analysis></analysis>. Write the answer in between <answer></answer>.
### Question:
A research group wants to assess the relationship between childhood diet and cardiovascular disease in adulthood. A prospective cohort study of 500 children between 10 to 15 years of age is conducted in which the participants' diets are recorded for 1 year and then the patients are assessed 20 years later for the presence of cardiovascular disease. A statistically significant association is found between childhood consumption of vegetables and decreased risk of hyperlipidemia and exercise tolerance. When these findings are submitted to a scientific journal, a peer reviewer comments that the researchers did not discuss the study's validity. Which of the following additional analyses would most likely address the concerns about this study's design??
{'A': 'Blinding', 'B': 'Crossover', 'C': 'Matching', 'D': 'Stratification', 'E': 'Randomization'},
### Response:
<analysis>
This is a question about assessing the validity of a prospective cohort study. The study found an association between childhood diet and cardiovascular disease in adulthood. The peer reviewer is concerned that the researchers did not discuss the validity of the study design.
To address concerns about validity in a prospective cohort study, we need to consider potential confounding factors that could influence the results. The additional analysis suggested should help control for confounding.4. Model Inference Before Fine-Tuning
Before fine-tuning the model, we will test its base performance to establish a baseline for comparison. This involves creating an inference prompt, selecting a sample question from the dataset, and generating a response using the base model.
We will create a prompt template for inference that includes a placeholder for the question.
inference_prompt_style = """
Please answer with one of the options in the bracket. Write reasoning in between <analysis></analysis>. Write the answer in between <answer></answer>.
### Question:
{}
### Response:
<analysis>
"""Then, we will select the question from the 11th sample and provide it to the model after formatting and converting it into tokens.
question = dataset[10]['input']
question = question.replace("Q:", "")
inputs = tokenizer(
[inference_prompt_style.format(question) + tokenizer.eos_token],
return_tensors="pt"
).to("cuda")
outputs = model.generate(
input_ids=inputs.input_ids,
attention_mask=inputs.attention_mask,
max_new_tokens=1200,
eos_token_id=tokenizer.eos_token_id,
use_cache=True,
)
response = tokenizer.batch_decode(outputs, skip_special_tokens=True)
print(response[0].split("### Response:")[1])The model's response was unsatisfactory. The analysis section was excessively long, consuming the token limit, and the answer section was never generated due to the lengthy reasoning.

5. Setting up the Model
In this step, we will set up the model and tokenizer for fine-tuning.
1. The SFTTrainer does not directly accept a tokenizer. Instead, we will convert the tokenizer into a data collator using the DataCollatorForLanguageModeling class from the transformers library.
from transformers import DataCollatorForLanguageModeling
data_collator = DataCollatorForLanguageModeling(
tokenizer=tokenizer,
mlm=False
)We will use LoRA (Low-Rank Adaptation) to fine-tune the model. LoRA modifies only a small subset of the model's parameters, making it highly memory-efficient and faster compared to full fine-tuning.
from peft import LoraConfig, get_peft_model
# LoRA config
peft_config = LoraConfig(
lora_alpha=16, # Scaling factor for LoRA
lora_dropout=0.05, # Add slight dropout for regularization
r=64, # Rank of the LoRA update matrices
bias="none", # No bias reparameterization
task_type="CAUSAL_LM", # Task type: Causal Language Modeling
target_modules=[
"q_proj",
"k_proj",
"v_proj",
"o_proj",
"gate_proj",
"up_proj",
"down_proj",
], # Target modules for LoRA
)
model = get_peft_model(model, peft_config)The TrainingArguments allows us to configure the fine-tuning process and the SFTTrainer simplifies the fine-tuning process by integrating the model, dataset, data collator, training arguments, and LoRA configuration into a single workflow.
from trl import SFTTrainer
from transformers import TrainingArguments
# Training Arguments
training_arguments = TrainingArguments(
output_dir="DeepSeek-R1-0528-Qwen3-8B-Medical-Reasoning",
per_device_train_batch_size=1,
per_device_eval_batch_size=1,
gradient_accumulation_steps=2,
optim="paged_adamw_32bit",
num_train_epochs=1,
logging_steps=0.2,
warmup_steps=10,
logging_strategy="steps",
learning_rate=2e-4,
fp16=False,
bf16=False,
group_by_length=True,
report_to="none"
)
# Initialize the Trainer
trainer = SFTTrainer(
model=model,
args=training_arguments,
train_dataset=dataset,
peft_config=peft_config,
data_collator=data_collator,
)6. Model Training
Before starting the training process, it is important to clear the cache and remove unwanted memory and VRAM footprints to avoid running into Out of Memory (OOM) errors.
Once the memory is cleared, we can start the training process using the SFTTrainer.
import gc, torch
gc.collect()
torch.cuda.empty_cache()
model.config.use_cache = False
trainer.train()During training, you can monitor the GPU by going to your RunPod dashboard. You should observe that the GPU utilization is close to 100%, which indicates that the model is being trained on the GPU (not the CPU).
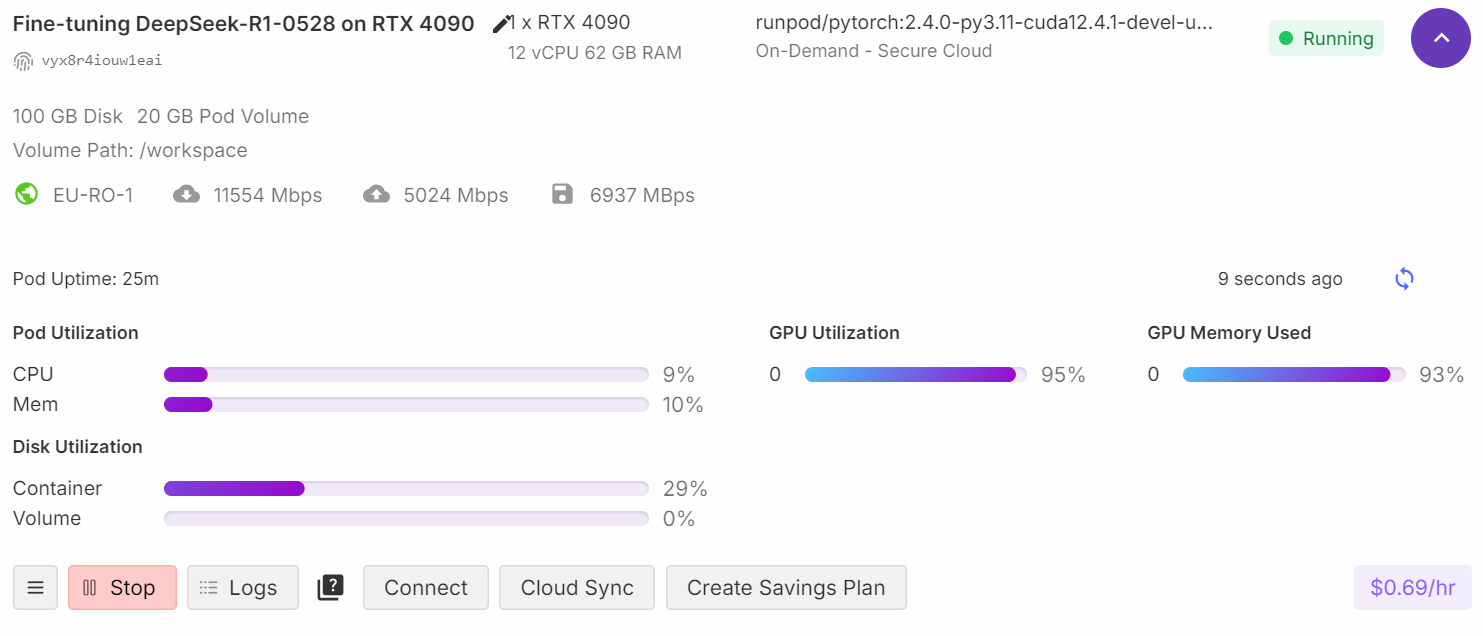
As the training progresses, you should notice that the training loss decreases gradually. This indicates that the model is learning and fine-tuning successfully.
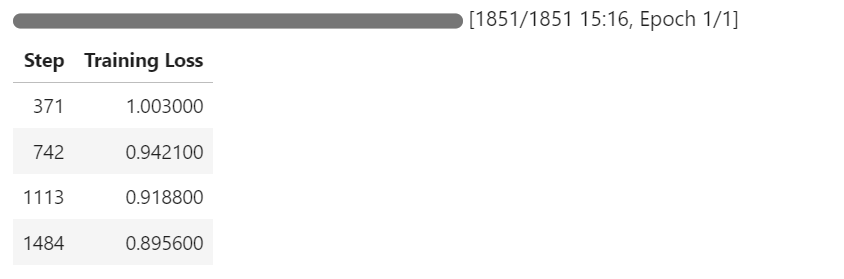
7. Model Inference After Fine-Tuning
After fine-tuning the model, we will test its performance by providing the same 11th sample question from the dataset. This allows us to compare the results with the base model and evaluate the improvements achieved through fine-tuning.
question = dataset[10]['input']
question = question.replace("Q:", "")
inputs = tokenizer(
[inference_prompt_style.format(question,) + tokenizer.eos_token],
return_tensors="pt"
).to("cuda")
outputs = model.generate(
input_ids=inputs.input_ids,
attention_mask=inputs.attention_mask,
max_new_tokens=1200,
eos_token_id=tokenizer.eos_token_id,
use_cache=True,
)
response = tokenizer.batch_decode(outputs, skip_special_tokens=True)
print(response[0].split("### Response:")[1])The fine-tuned model's response shows significant improvements:
- The analysis section is concise and uses the <analysis> tag correctly instead of <think>.
- The answer section is formatted correctly within the <answer> tag.
- However, the answer itself is incorrect, as the model selected E: Randomization, which does not match the correct answer in the dataset.
<analysis>
This is a question about evaluating the validity of a prospective cohort study design. The study looked at childhood diet and cardiovascular disease in adulthood. The peer reviewer raised concerns about the study's validity, likely because it was an observational study without randomization or control group.
The question asks which additional analysis would most likely address the concerns about the study's design. The choices include blinding, crossover, matching, stratification, and randomization.
Randomization is the key to reducing bias in observational studies. By randomly assigning participants to different groups, you can control for confounding factors. The other choices do not directly address the lack of randomization in the original study design.
</analysis>
<answer>
E: Randomization
</answer>To further evaluate the fine-tuned model, we will test it on another sample from the dataset and observe the results.
question = dataset[100]['input']
question = question.replace("Q:", "")
inputs = tokenizer(
[inference_prompt_style.format(question) + tokenizer.eos_token],
return_tensors="pt"
).to("cuda")
outputs = model.generate(
input_ids=inputs.input_ids,
attention_mask=inputs.attention_mask,
max_new_tokens=1200,
eos_token_id=tokenizer.eos_token_id,
use_cache=True,
)
response = tokenizer.batch_decode(outputs, skip_special_tokens=True)
print(response[0].split("### Response:")[1])The fine-tuned model performs perfectly on this sample:
- The analysis section is concise, accurate, and matches the dataset's style.
- The answer section is correct and formatted properly.
<analysis>
This is a clinical vignette describing a 55-year-old man with burning and shooting pain in his feet and lower legs that worsens at night. He has a history of type 2 diabetes mellitus and hypertension. The description of the pain being burning and shooting in a symmetric distribution in the lower extremities, worsening at night, along with his history of diabetes, is most consistent with distal symmetric sensorimotor polyneuropathy. Autonomic neuropathy would not cause sensory symptoms. Isolated cranial nerve or peripheral nerve neuropathy would not explain the symmetric distribution. Radiculopathy would not explain the sensory symptoms.
</analysis>
<answer>
D: Distal symmetric sensorimotor polyneuropathy
</answer>8. Saving the Model
The next step is to save the fine-tuned model and push it to the Hugging Face Hub. This allows others to access and use the fine-tuned model for inference or further fine-tuning.
new_model_name = "DeepSeek-R1-0528-Qwen3-8B-Medical-Reasoning"
trainer.model.push_to_hub(new_model_name)
trainer.processing_class.push_to_hub(new_model_name)Once the model is successfully pushed, it will be available on the Hugging Face Hub: kingabzpro/DeepSeek-R1-0528-Qwen3-8B-Medical-Reasoning.
9. Loading the Fine-Tuned Model From Hugging Face
After saving the fine-tuned model and adapter to the Hugging Face Hub, we will now test if the model performs as expected by loading it back. This involves clearing the GPU memory, loading the base model and LoRA adapter, and running inference on a sample prompt.
1. Clear the GPU memory to avoid any potential Out of Memory (OOM) issues. This ensures a clean environment for loading the model.
del model
del trainer
torch.cuda.empty_cache()2. load the base model and the fine-tuned LoRA adapter from the Hugging Face Hub. The base model will be loaded in 4-bit quantization to optimize memory usage, and the LoRA adapter will be merged with it.
from transformers import AutoTokenizer, AutoModelForCausalLM, BitsAndBytesConfig
from peft import PeftModel
import torch
# Base model
base_model_id = "deepseek-ai/DeepSeek-R1-0528-Qwen3-8B"
# Your fine-tuned LoRA adapter repository
lora_adapter_id = "kingabzpro/DeepSeek-R1-0528-Qwen3-8B-Medical-Reasoning"
# Load the model in 4-bit
bnb_config = BitsAndBytesConfig(
load_in_4bit=True,
bnb_4bit_use_double_quant=False,
bnb_4bit_quant_type="nf4",
bnb_4bit_compute_dtype=torch.bfloat16,
)
# Load base model
base_model = AutoModelForCausalLM.from_pretrained(
base_model_id,
device_map="auto",
torch_dtype=torch.bfloat16,
quantization_config=bnb_config,
trust_remote_code=True,
)
# Attach the LoRA adapter
model = PeftModel.from_pretrained(
base_model,
lora_adapter_id,
device_map="auto",
trust_remote_code=True,
)
# Load tokenizer
tokenizer = AutoTokenizer.from_pretrained(base_model_id, trust_remote_code=True)3. We will now test the fine-tuned model by providing a sample prompt. The prompt will be tokenized, passed through the model, and the response will be decoded.
# Inference example
prompt = """
Please answer with one of the options in the bracket. Write reasoning in between <analysis></analysis>. Write the answer in between <answer></answer>.
### Question:
A research group wants to assess the relationship between childhood diet and cardiovascular disease in adulthood.
A prospective cohort study of 500 children between 10 to 15 years of age is conducted in which the participants' diets are recorded for 1 year and then the patients are assessed 20 years later for the presence of cardiovascular disease.
A statistically significant association is found between childhood consumption of vegetables and decreased risk of hyperlipidemia and exercise tolerance.
When these findings are submitted to a scientific journal, a peer reviewer comments that the researchers did not discuss the study's validity.
Which of the following additional analyses would most likely address the concerns about this study's design?
{'A': 'Blinding', 'B': 'Crossover', 'C': 'Matching', 'D': 'Stratification', 'E': 'Randomization'},
### Response:
<analysis>
"""
inputs = tokenizer(
[prompt + tokenizer.eos_token],
return_tensors="pt"
).to("cuda")
outputs = model.generate(
input_ids=inputs.input_ids,
attention_mask=inputs.attention_mask,
max_new_tokens=1200,
eos_token_id=tokenizer.eos_token_id,
use_cache=True,
)
response = tokenizer.batch_decode(outputs, skip_special_tokens=True)
print(response[0].split("### Response:")[1])The fine-tuned model produces a response similar to the dataset, which confirms that the model and adapter were loaded correctly.
<analysis>
This is a question about evaluating the validity of a prospective cohort study design. The study looked at childhood diet and cardiovascular disease in adulthood. The peer reviewer was concerned about the study's validity.
To address concerns about validity in a prospective cohort study, we need to consider potential confounding factors. The choices given are different statistical methods that can help control for confounding.
Blinding and crossover designs are not applicable to a prospective cohort study. Matching and stratification can help control for confounding by balancing the distribution of confounders between groups. Randomization is the best way to minimize confounding by randomly assigning participants to different exposure groups.
</analysis>
<answer>
E: Randomization
</answer>If you encounter any issues while running the above code, refer to the fine-tuning-Deepseek-new-R1.ipynb notebook. This notebook has been tested multiple times and should work out of the box when loaded into RunPod or a similar environment.
Conclusion
The DeepSeek-R1-0528 model is one of the best open-source reasoning models available today. Its ability to perform complex reasoning tasks, combined with its open-source nature, makes it a standout choice for developers and researchers. While the fine-tuned model already delivers impressive results, there are several ways to further improve fine-tuning outcomes:
- Load the full model: Using the full model instead of quantized versions can improve accuracy.
- Train for more epochs: Training the model for at least 3 epochs instead of 1 can lead to better convergence and improved performance.
- Refine the prompt style: Experimenting with and optimizing the prompt style can help the model better understand and respond to specific tasks.
- Optimize hyperparameters: Training hyperparameters such as learning rate, batch size, and gradient accumulation steps can significantly enhance results.
- Increase dataset size: Training on a larger dataset (e.g., more than 10,000 samples) can improve the model's generalization and reasoning capabilities.
Moreover, this guide can be used to fine-tune the DeepSeek R1 model on any reasoning dataset, including synthetic datasets created using other models like OpenAI's o3. You can also get hands-on with some of the techniques shown here in our Fine-Tuning With Llama 3 course. If you're interested in fine-tuning other models with medical datasets, check out our tutorial, Fine-Tuning MedGemma on a Brain MRI Dataset.

As a certified data scientist, I am passionate about leveraging cutting-edge technology to create innovative machine learning applications. With a strong background in speech recognition, data analysis and reporting, MLOps, conversational AI, and NLP, I have honed my skills in developing intelligent systems that can make a real impact. In addition to my technical expertise, I am also a skilled communicator with a talent for distilling complex concepts into clear and concise language. As a result, I have become a sought-after blogger on data science, sharing my insights and experiences with a growing community of fellow data professionals. Currently, I am focusing on content creation and editing, working with large language models to develop powerful and engaging content that can help businesses and individuals alike make the most of their data.


