Course
Self-organizing maps (SOMs) are a type of artificial neural network used for unsupervised learning tasks such as clustering. Given a set of data points, the model learns to divide the data into clusters. SOMs project complex multidimensional data onto a lower-dimensional (typically, 2-dimensional) grid. This facilitates easier visualization. Hence, SOMs are also used to visualize high-dimensional data.
In this tutorial, we explore the core concepts of SOMs, their learning process, and their typical use cases. We explain how to implement a SOM in Python using the MiniSom library and how to represent the results visually. Finally, we discuss important hyperparameters in training SOM models and how to fine-tune them.
Understanding Self-Organizing Maps (SOM)
In this section, we present the core concepts of SOMs, their learning process, and their use cases.
Core concepts of SOMs
Self-organizing maps involve a set of concepts that are important to understand.
Neurons
A SOM is essentially a grid of neurons. During training, the neuron whose weight vector is closest to an input data point adjusts its weights and neighbors’ weights to match the input data point even more closely. Over many iterations, groups of neighboring neurons map to related data points. This results in the clustering of the input dataset.
Grid structure
The neurons in a SOM are organized as a grid. Neighboring neurons map to similar data points. Larger datasets need a larger grid. Typically, this grid is two-dimensional. The grid structure serves as the low-dimensional space onto which high-dimensional data is mapped. The grid aids in visualizing data patterns and clusters.
Mapping data points
Each data point is compared to all the neurons using a distance metric. The neuron whose weight vector is closest to the input data point is the Best Matching Unit (BMU) for that data point.
Once the BMU is identified, the weights of the BMU and its neighboring neurons are updated. This update brings the BMU and the neighboring neurons even closer to the input data point. This mapping preserves the data’s topology, ensuring that similar data points are mapped to nearby neurons.
Learning process of SOMs
SOM training is considered unsupervised because it is not based on labeled datasets. The goal of training SOMs is to iteratively adjust the neurons' weight vectors so that similar data points are mapped to nearby neurons.
Competitive learning
SOMs use competitive learning (instead of gradient descent and backpropagation). Neurons compete to become the Best Matching Unit (BMU) for each input data point. The neuron closest to a data point is determined to be its BMU. The BMU and its neighboring neurons get updated to further decrease their distance to the data point. Neighboring neurons map to related data points. This leads to specialization among the neurons and clustering of the input data points.
Distance functions
SOMs use a distance function to measure the distance between neurons and data points. This distance is used to determine the BMU of each data point. MiniSom has four distance functions to choose from:
- Euclidean distance (this is the default choice)
- Cosine distance
- Manhattan distance
- Chebyshev distance
Neighborhood function
After identifying the BMU of a data point, the BMU and its neighboring neurons are updated in the direction of that data point. The neighborhood function ensures the SOM maintains the topological relationships of the input data. The neighborhood function decides:
- Which neurons are considered to be in the BMU’s neighborhood
- The extent to which the neighboring neurons are updated. In general, neurons closer to the BMU receive a larger adjustment than those further away
MiniSom comes with three neighborhood functions:
- Gaussian (this is the default).
- Bubble
- Mexican hat
- Triangle
Iterative process
The learning process in SOMs happens over several iterations. In each iteration, the SOM processes many input data points. We outline the learning process in the following steps:
- When the training starts, the weights of all neurons are (randomly) initialized.
- Given an input data point, each neuron calculates its distance from the input.
- The neuron with the smallest distance is declared the BMU.
- The weights of the BMU and its neighbors are adjusted to move closer to the input vector. The extent of this adjustment is decided by:
- A neighborhood function: Neurons farther away from the BMU are updated less than those closer to the BMU. This preserves the topological structure of the data.
- Learning rate: A higher learning rate leads to larger updates.
These steps are repeated over many iterations, going over many input data points. The weights are gradually updated so that the map self-organizes and captures the structure of the data.
The learning rate and neighborhood radius typically decrease over time, allowing the SOM to fine-tune the weights gradually. Later in the tutorial, we implement the steps of the iterative training process using Python code.
Typical use cases of SOMs
Given their unique architecture, SOMs have several typical applications in machine learning, analysis, and visualization.
Clustering
SOMs are used to group high-dimensional data into clusters of similar data points. This helps to identify inherent structures within the data. Each neuron maps to closely related data points and similar data points are mapped to the same or neighboring neurons, thus forming distinct clusters.
For example, in marketing, SOMs can group customers based on purchasing behavior. This would allow a brand to tailor marketing strategies to different customer segments (clusters).
Dimensionality reduction
SOMs can map high-dimensional data with many feature vectors to a grid with fewer dimensions (typically, a 2-dimensional grid). Since this mapping preserves the relationships between data points, the reduced dimensionality makes it easier to visualize complex data sets without significant loss of information.
In addition to making visualization and analysis tasks easier, SOMs can also reduce the dimensionality of data before applying other machine learning algorithms. SOMs are also used in image processing to reduce the number of features by clustering pixels or regions with similar characteristics, making image recognition and classification tasks more efficient.
Anomaly detection
During the training process, most data points are mapped to specific neurons. Anomalies (data points that don’t fit well into any cluster) are typically mapped to distant or less populated neurons. These outliers do not fit well into the learned clusters and are mapped to neurons far from the BMU of normal data points.
For example, mapping financial transactions on a SOM grid allows us to identify unusual activities (outliers) that can indicate potentially fraudulent activity.
Data visualization
Data visualization is one of the most common use cases of SOMs. High-dimensional data is difficult to visualize. SOMs make it easier to visualize by reducing the dimensionality of the data and projecting it on a two-dimensional grid. This can also help detect clusters in the data and anomalous data points that would be complicated to discover in the original data.
For example, data visualization makes it easier to analyze high-dimensional population data. Mapping the data on a 2D grid would help spot similarities and differences between different population groups.
After discussing the core concepts and use cases of SOMs, we will show how to implement them using the Python package MiniSom in the following sections.
You can access and run the complete code on this DataLab notebook.
Build Machine Learning Skills
Setting Up the Environment for SOM
Before building the SOM, we need to prepare the environment with the necessary packages.
Installing Python libraries
We need these packages:
- MiniSom is a NumPy-based Python tool that creates and trains SOMs.
- NumPy is used to access mathematical functions such as splitting arrays, getting unique values, etc.
matplotlibis used to plot various graphs and charts to visualize the data.- The
datasetspackage fromsklearnis used to import datasets on which to apply the SOM. - The
MinMaxScalerpackage fromsklearnnormalizes the dataset.
The following code snippet imports these packages:
from minisom import MiniSom
import numpy as np
import matplotlib.pyplot as plt
from sklearn import datasets
from sklearn.preprocessing import MinMaxScalerPreparing the dataset
In this tutorial, we use MiniSom to build a SOM and then train it on the canonical IRIS dataset. This dataset consists of 3 classes of iris plants. Each class has 50 instances. To prepare the data, we follow these steps:
- Import the Iris dataset from
sklearn, - Extract the data vectors and the target scalars.
- Normalize the data vectors. In this tutorial, we use the MinMaxScaler from scikit-learn.
- Declare a set of labels for each of the three classes of Iris plants.
The following code implements these steps:
dataset_iris = datasets.load_iris()
data_iris = dataset_iris.data
target_iris = dataset_iris.target
data_iris_normalized = MinMaxScaler().fit_transform(data_iris)
labels_iris = {1:'1', 2:'2', 3:'3'}
data = data_iris_normalized
target = target_irisImplementing Self-Organizing Maps (SOM) in Python
To implement a SOM in Python, we define and initialize the grid before training it on the dataset. We can then visualize the trained neurons and the clustered dataset.
Defining the SOM grid
As explained earlier, a SOM is a grid of neurons. Using MiniSom, we can create 2-dimensional grids. The grid’s X and Y dimensions are the number of neurons along each axis. To define the SOM grid, we also need to specify:
- The X and Y dimensions of the grid
- The number of input variables - this is the number of data rows.
Declare these parameters as Python constants:
SOM_X_AXIS_NODES = 8
SOM_Y_AXIS_NODES = 8
SOM_N_VARIABLES = data.shape[1]The sample code below illustrates how to declare the grid using MiniSom:
som = MiniSom(SOM_X_AXIS_NODES, SOM_Y_AXIS_NODES, SOM_N_VARIABLES)The first two parameters are the number of neurons along the X and Y axes, and the third parameter is the number of variables.
We declare other parameters and hyperparameters while creating the SOM grid. We will explain these later in the tutorial. For now, declare these parameters as shown below:
ALPHA = 0.5
DECAY_FUNC = 'linear_decay_to_zero'
SIGMA0 = 1.5
SIGMA_DECAY_FUNC = 'linear_decay_to_one'
NEIGHBORHOOD_FUNC = 'triangle'
DISTANCE_FUNC = 'euclidean'
TOPOLOGY = 'rectangular'
RANDOM_SEED = 123Create a SOM using these parameters:
som = MiniSom(
SOM_X_AXIS_NODES,
SOM_Y_AXIS_NODES,
SOM_N_VARIABLES,
sigma=SIGMA0,
learning_rate=ALPHA,
neighborhood_function=NEIGHBORHOOD_FUNC,
activation_distance=DISTANCE_FUNC,
topology=TOPOLOGY,
sigma_decay_function = SIGMA_DECAY_FUNC,
decay_function = DECAY_FUNC,
random_seed=RANDOM_SEED,
)Initializing the neurons
The above command creates a SOM with random weights for all the neurons. Initializing the neurons with weights drawn from the data (instead of random numbers) can make the training process more efficient.
When using MiniSom to create a Self-Organizing Map (SOM), there are two ways to initialize the neurons’ weights based on the data:
- Random initialization: The neurons' initial weights are randomly drawn from the input data. We do this by applying the
.random_weights_init()function to the SOM. - PCA initialization: Principal Component Analysis (PCA) initialization uses the principal components of the input data to initialize the weights. The initial weights of the neurons span the first two principal components. This often leads to faster convergence.
In this guide, we use PCA initialization. To apply PCA initialization on the SOM weights, use the .pca_weights_init() function as shown below:
som.pca_weights_init(data)Training the SOM
The training process updates the SOM weights to minimize the distance between the neurons and the data points.
Below, we explain the iterative training process:
- Initialization: The weight vectors of all the neurons are initialized, typically with random values. It is also possible to initialize the weights by sampling the input data distribution.
- Input selection: An input vector is (randomly) selected from the training dataset.
- BMU identification: The neuron with the weight vector closest to the input vector is identified as the BMU.
- Neighborhood update: The BMU and its neighboring neurons update their weight vectors. The learning rate and the neighborhood function decide which neurons are updated and by how much. At the iteration step t, given the input vector x, the weight vector of neuron i as wi, the learning rate (t), and the neighborhood function hbi (this function quantifies the extent of update for the neuron i given the BMU neuron b), the weight update formula for neuron i is expressed as:

- Decay rate of learning rate and neighborhood radius: Both the learning rate and the neighborhood radius decrease over time. In the earlier iterations, the training process makes larger adjustments over a larger neighborhood. Later iterations help to fine-tune the weights by making smaller changes to the weights of adjacent neurons. This allows the map to stabilize and converge.
To train the SOM, we present the model with the input data. We can choose from one of two approaches to do this:
- Pick samples at random from the input data. The
.train_random()function implements this technique. - Sequentially run through the vectors in the input data. This is done using the
.train_batch()function.
These functions accept the input data and the number of iterations as parameters. In this guide, we use the .train_random() function. Declare the number of iterations as a constant and pass it to the training function:
N_ITERATIONS = 5000
som.train_random(data, N_ITERATIONS, verbose=True) After executing the script and completing the training, a message with the quantization error is displayed:
quantization error: 0.05357240680504421 The quantization error indicates the amount of information lost when the SOM quantizes (reduces the dimensionality of) the data. A large quantization error indicates a larger distance between the neurons and the data points. It also means that the clustering is less reliable.
Visualizing SOM neurons
We now have a trained SOM model. To visualize it, we use a distance map (also known as a U-matrix). The distance map displays the SOM’s neurons as a grid of cells. The color of each cell represents its distance from the neighboring neurons.
The distance map is a grid with the same dimensions as the SOM. Each cell in the distance map is the normalized sum of the (Euclidean) distances between a neuron and its neighbors.
Access the SOM distance map using the .distance_map() function. To generate the U-matrix, we follow these steps:
- Use
pyplotto create a figure with the same dimensions as the SOM. In this example, the dimensions are 8x8. - Plot the distance map using matplotlib using the
.pcolor()function. In this example, we usegist_yargas the color scheme. - Display the
colorbar, an index mapping different colors to different scalar values. In this case, since the distances are normalized, the scalar distance values range from 0 to 1.
The code below implements these steps:
# create the grid
plt.figure(figsize=(8, 8))
#plot the distance map
plt.pcolor(som.distance_map().T, cmap='gist_yarg')
# show the color bar
plt.colorbar()
plt.show()In this example, the U-matrix uses a monotone color scheme. It can be understood using these guidelines:
- Lighter shades represent closely spaced neurons, and darker shades represent neurons farther away from others.
- Groups of lighter shades can be interpreted as clusters. Dark nodes between the clusters can be interpreted as the boundaries between clusters.
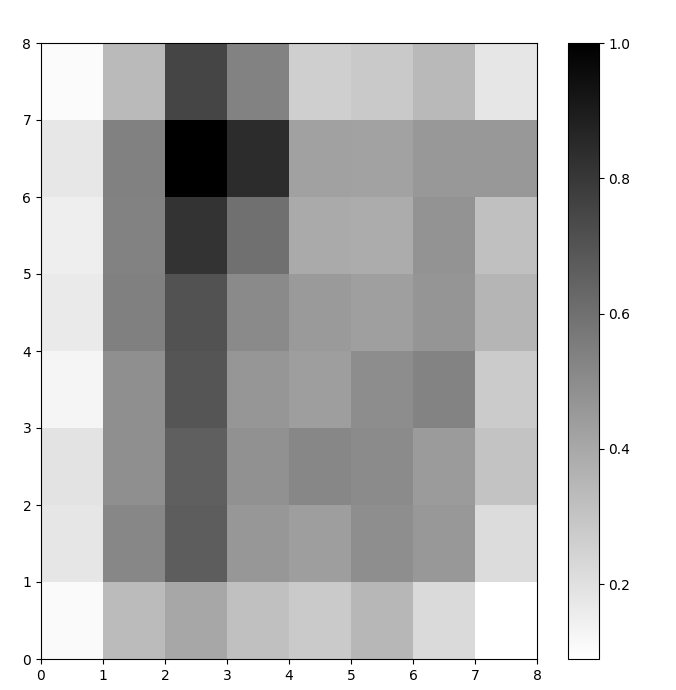
Figure 1: U-matrix of SOM trained on the Iris dataset (image by author)
Evaluating the SOM Clustering Results
The previous figure graphically illustrated the SOM’s neurons. In this section, we show how to visualize how the SOM clustered the data.
Identifying clusters
We overlay markers over the above U-matrix to denote which class of Iris plant each cell (neuron) represents. To do this:
- As before, create an 8x8 figure using
pyplot, plot the distance map, and show the color bar. - Specify an array of three matplotlib markers, one for each class of Iris plant.
- Specify an array of three matplotlib color codes, one for each class of Iris plant.
- Iteratively plot the winning neuron for each data point:
- Determine the (coordinates of the) winning neuron for each data point using the
.winner() function. - Plot the position of each winning neuron in the middle of each cell on the grid.
w[0]andw[1]give the neuron’s X and Y coordinates, respectively. A value of 0.5 is added to each coordinate to plot it in the middle of the cell.
The code below shows how to do this:
# plot the distance map
plt.figure(figsize=(8, 8))
plt.pcolor(som.distance_map().T, cmap='gist_yarg')
plt.colorbar()
# create the markers and colors for each class
markers = ['o', 'x', '^']
colors = ['C0', 'C1', 'C2']
# plot the winning neuron for each data point
for count, datapoint in enumerate(data):
# get the winner
w = som.winner(datapoint)
# place a marker on the winning position for the sample data point
plt.plot(w[0]+.5, w[1]+.5, markers[target[count]-1], markerfacecolor='None',
markeredgecolor=colors[target[count]-1], markersize=12, markeredgewidth=2)
plt.show()The resulting image is shown below:
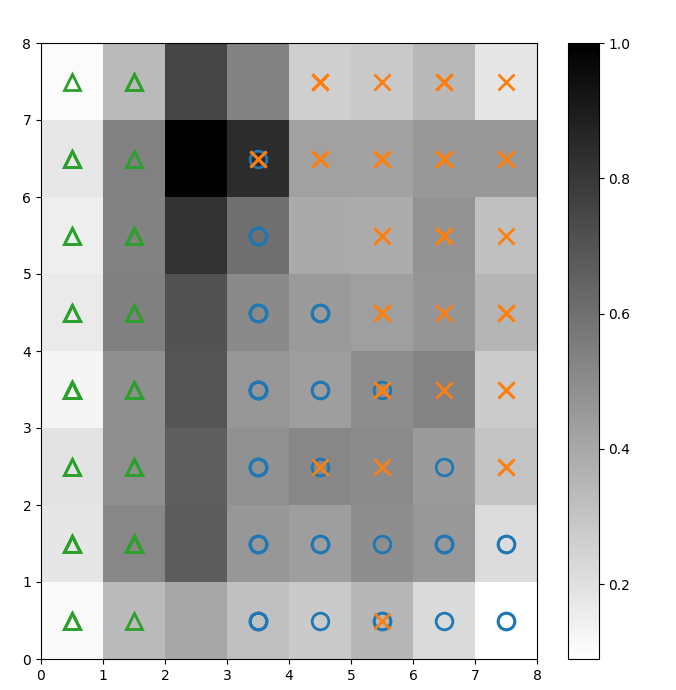
Figure 2: U-matrix overlaid with class markers (image by author)
Based on the Iris dataset documentation, “one class is linearly separable from the other 2; the latter are not linearly separable from each other”. In the U-matrix above, these three classes are represented by three markers - triangle, circle, and cross.
Notice that there isn’t a clear boundary between the blue circles and the orange crosses. Furthermore, two classes are overlaid on the same neuron in many cells. This means the neuron is equidistant from both classes.
Visualizing the clustering outcome
A SOM is a clustering model. Similar data points map to the same neuron. Datapoints of the same class map to a cluster of neighboring neurons. We plot all the data points on the SOM grid to better study the clustering behavior.
The following steps describe how to create this scatter plot:
- Get the winning neuron’s X and Y coordinates of the winning neuron for each data point.
- Plot the distance map, as we did for Figure 1.
- Use
plt.scatter()to make a scatter plot of all the winning neurons for each data point. Add a random offset to each point to avoid overlaps between data points within the same cell.
We implement these steps in the code below:
# get the X and Y coordinates of the winning neuron for each data pointw_x, w_y = zip(*[som.winner(d) for d in data])
w_x = np.array(w_x)
w_y = np.array(w_y)
# plot the distance map
plt.figure(figsize=(8, 8))
plt.pcolor(som.distance_map().T, cmap='gist_yarg', alpha=.2)
plt.colorbar()
# make a scatter plot of all the winning neurons for each data point
# add a random offset to each point to avoid overlaps
for c in np.unique(target):
idx_target = target==c
plt.scatter(w_x[idx_target]+.5+(np.random.rand(np.sum(idx_target))-.5)*.8,
w_y[idx_target]+.5+(np.random.rand(np.sum(idx_target))-.5)*.8,
s=50,
c=colors[c-1],
label=labels_iris[c+1]
)
plt.legend(loc='upper right')
plt.grid()
plt.show()The following graph shows the output scatter plot:
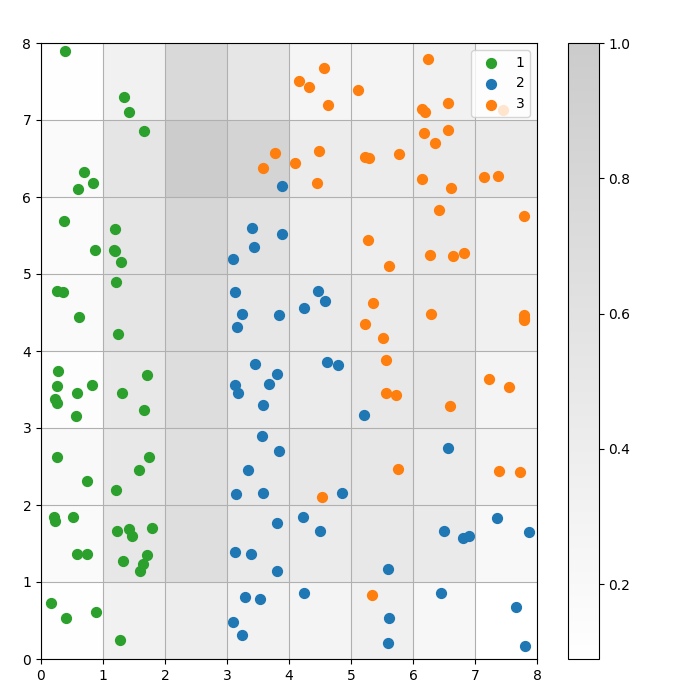 Figure 3: Scatter plot of data points within cells (image by author)
Figure 3: Scatter plot of data points within cells (image by author)
In the above scatter plot, observe that:
- Some cells contain both blue and orange dots.
- The green dots are clearly separated from the rest of the data, but the blue and orange dots are not cleanly separated.
- The above observations align with the fact that only one of the three clusters in the Iris dataset has a clear boundary.
- In Figure 1, dark nodes between the clusters (which can be interpreted as the boundaries between clusters) match with empty cells in the scatter plot.
You can access and run the complete code on this DataLab notebook.
Tuning the SOM Model
The previous sections showed how to create and train a SOM model and how to study the results visually. In this section, we discuss how to tune the performance of SOM models.
Key hyperparameters to tune
As with any machine learning model, hyperparameters considerably impact the model’s performance.
Some of the hyperparameters important in training SOMs are:
- The grid size decides the map’s size. The number of neurons in a map with a grid size of AxB is A*B.
- The learning rate decides how much the weights are changed in each iteration. We set the initial learning rate, and it decreases over time according to the decay function.
- The decay function decides the extent to which the learning rate is decreased in each subsequent iteration.
- The neighborhood function is a mathematical function that specifies which neurons are to be considered as the BMU’s neighbors.
- The standard deviation specifies the neighborhood function’s spread. For example, a Gaussian neighborhood function with a high standard deviation will have a larger neighborhood than the same function with a smaller standard deviation. We set the initial standard deviation, which decreases over time according to the sigma decay function.
- The sigma decay function controls how much the standard deviation is reduced in each subsequent iteration.
- The number of training iterations decides how many times the weights are updated. In each training iteration, the neuron weights are updated once.
- The distance function is a mathematical function that calculates the distance between neurons and data points.
- The topology decides the grid structure’s layout. The neurons in the grid can be arranged in a rectangular or hexagonal pattern.
In the next section, we discuss guidelines for setting the values of these hyperparameters.
Impact of hyperparameter tuning
Hyperparameter values should be decided based on the model and the dataset. To some extent, determining these values is a process of trial and error. In this section, we give guidelines for tuning each hyperparameter. Next to each hyperparameter, we mention (in parentheses) the respective Python constants used in the sample code.
- Grid size (
SOM_X_AXIS_NODESandSOM_Y_AXIS_NODES): The grid size depends on the dataset’s size. The rule of thumb is that given a dataset of size N, the grid should contain roughly 5*sqrt(N) neurons. For example, if the dataset has 150 samples, the grid should contain 5*sqrt(150) = approximately 61 neurons. In this tutorial, the Iris dataset has 150 rows and we use an 8x8 grid. - Initial learning rate (
ALPHA): A higher rate speeds up convergence, while lower rates are used for finer adjustments after early iterations. The initial learning rate should be large enough to enable quick adaptation but not so large that it overshoots optimal weight values. In this article, the initial learning rate is 0.5. - Initial standard deviation (
SIGMA0): It determines the neighborhood’s initial size or spread. A larger value considers more global patterns. In this example, we use a starting standard deviation of 1.5. - For the decay rate (
DECAY_FUNC) and the sigma decay rate (SIGMA_DECAY_FUNC), we can choose from one of three types of decay functions: - Inverse decay: This function is suitable if the data has both global and local patterns. In such cases, we need a longer phase of broad learning before focusing on local patterns.
- Linear decay: This is good for datasets where we want a steady and uniform neighborhood size or learning rate reduction. This is useful if the data doesn't need much fine-tuning.
- Asymptotic decay: This function is useful if the data is complex and high-dimensional. In such cases, it is better to spend more time on global exploration before gradually transitioning to finer details.
- Neighborhood function (
NEIGHBORHOOD_FUNC): The default choice of the neighborhood function is the Gaussian function. Other functions, as explained below, are also used. - Gaussian (default): This is a bell-shaped curve. The extent to which a neuron is updated decreases smoothly as its distance from the winning neuron increases. It provides a smooth and continuous transition and preserves the data’s topology. It is suitable for most general purposes due to its stable and predictable behavior.
- Bubble: This function creates a fixed-width neighborhood. All neurons within this neighborhood are updated equally, and neurons outside this neighborhood are not updated (for a given data point). It is computationally cheaper and easier to implement. It is useful for smaller maps where sharp neighborhood boundaries do not compromise effective clustering.
- Mexican hat: It has a central positive region surrounded by a negative region. Neurons close to the BMU are updated to come closer to the data point, and neurons further away are updated to move away from the data point. This technique enhances contrast and sharpens the features in the map. Since it emphasizes distinct clusters, it is effective in pattern recognition tasks where a clear separation of clusters is desired.
- Triangle: This function defines the neighborhood size as a triangle, with the BMU having the largest influence. It decreases linearly with distance from the BMU. It is used for clustering data with gradual transitions between clusters or features, such as image, speech, or time-series data, where neighboring data points are expected to share similar characteristics.
- Distance function (
DISTANCE_FUNC): To measure the distance between neurons and data points, we can choose from 4 methods: - Euclidean distance (default choice): Useful when the data is continuous, and we want to measure straight-line distance. It suits most general tasks, especially when data points are evenly distributed and spatially related.
- Cosine distance: Good choice for text or high-dimensional sparse data where the angle between vectors is more important than magnitude. It’s useful for comparing directionality in data.
- Manhattan distance: Ideal when data points are on a grid or lattice (e.g., city blocks). This is less sensitive to outliers than Euclidean distance.
- Chebyshev distance: Suitable for situations where movement can occur in any direction (e.g., chessboard distances). It is useful for discrete spaces where we want to prioritize the maximum axis difference.
- Topology (
TOPOLOGY): In a grid, neurons can be arranged in a hexagonal or rectangular structure: - Rectangular (default): each neuron has 4 immediate neighbors. This is the right choice when the data doesn’t have a clear spatial relationship. It is also computationally simpler.
- Hexagonal: each neuron has 6 neighbors. This is the preferred option if the data has spatial relationships better represented with a hexagonal grid. This is the case for circular or angular data distributions.
- Number of training iterations (
N_ITERATIONS): In principle, longer training times lead to lower errors and better alignment of the weights with the input data. However, the model’s performance increases asymptotically with the number of iterations. Thus, after a certain number of iterations, the performance increase from subsequent interactions is only marginal. Deciding the correct number of iterations takes some experimentation. In this tutorial, we train the model over 5000 iterations.
To determine the right configuration of hyperparameters, we recommend experimenting with various options on a smaller subset of the data.
Conclusion
Self-organizing maps are a robust tool for unsupervised learning. They are used for clustering, dimensionality reduction, anomaly detection, and data visualization. Since they preserve the topological properties of high-dimensional data and represent it on a lower-dimensional grid, SOMs make it easy to visualize and interpret complex datasets.
This tutorial discussed the underlying principles of SOMs and showed how to implement a SOM using the MiniSom Python library. It also demonstrated how to visually analyze the results and explained the important hyperparameters used to train SOMs and finetune their performance.
Become an ML Scientist
FAQs
Does SOM training involve backpropagation?
A SOM is an artificial neural network, but its training is not based on error correction. Thus, it does not use backpropagation with gradient descent. Instead, SOMs are trained using competitive learning.
What is the need for the decay functions?
Early in the training process, the model needs to explore the global landscape of the data. Thus, the learning rate needs to be high, and the neighborhood function needs to be spread widely. Later training iterations make finer adjustments to the weights based on adjacent neurons. As training progresses, the decay functions reduce the learning rate and the neighborhood radius.
What is the difference between the default (random) initialization and random initialization based on the input data?
In the default initialization, the weights are initialized to arbitrary random numbers.
In random initialization based on the input data, the weights are initialized to randomly chosen samples from the input data.
Can SOMs capture non-linear relationships in the data?
Yes, SOMs capture non-linear patterns in the data by using various neighborhood functions. A single data point can affect the values of many neurons (in the neighborhood). This allows SOMs to capture complex relationships in the data.
What is the difference between SOM and traditional clustering methods?
SOMs map high-dimensional data to a lower-dimensional grid, preserving spatial relationships. On the other hand, traditional clustering techniques like K-Means create groups based on distance metrics without necessarily accounting for data topology or spatial relationships.
Note that in data analysis, topological or spatial relationships refer to how data points are positioned relative to each other in a multi-dimensional space.
Arun is a former startup founder who enjoys building new things. He is currently exploring the technical and mathematical foundations of Artificial Intelligence. He loves sharing what he has learned, so he writes about it.
In addition to DataCamp, you can read his publications on Medium, Airbyte, and Vultr.




